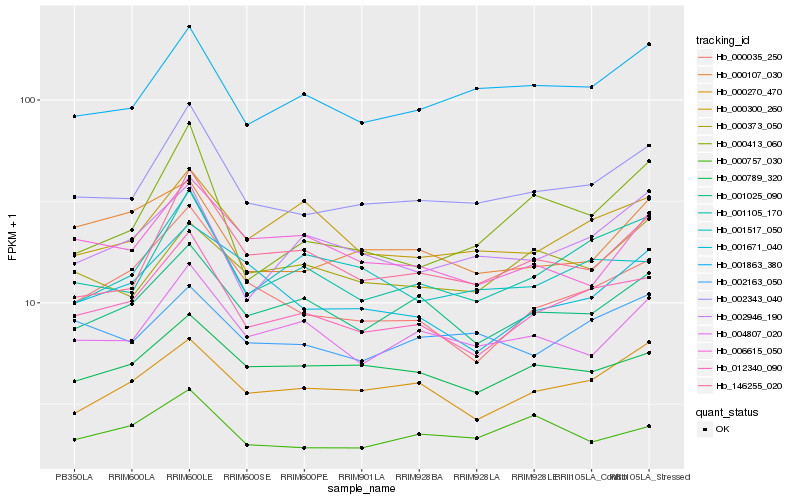
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002343_040 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001863_380 |
0.0702245015 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 3 |
Hb_012340_090 |
0.0704000379 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 4 |
Hb_001105_170 |
0.0735048328 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 5 |
Hb_146255_020 |
0.0790357113 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 6 |
Hb_004807_020 |
0.0797679689 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 7 |
Hb_001025_090 |
0.0857418908 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 8 |
Hb_000789_320 |
0.0892008472 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002163_050 |
0.089588567 |
- |
- |
hypothetical protein POPTR_0014s15490g [Populus trichocarpa] |
| 10 |
Hb_000270_470 |
0.0935616816 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: pentatricopeptide repeat-containing protein At1g74900, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000757_030 |
0.0948618614 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 12 |
Hb_002946_190 |
0.096766798 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001517_050 |
0.0989324882 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000373_050 |
0.0999021137 |
- |
- |
PREDICTED: protein WHI3 [Jatropha curcas] |
| 15 |
Hb_006615_050 |
0.100058899 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 16 |
Hb_000107_030 |
0.100370765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000413_060 |
0.10100535 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 18 |
Hb_000035_250 |
0.1012705361 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 19 |
Hb_000300_260 |
0.1034753455 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 20 |
Hb_001671_040 |
0.103658291 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |