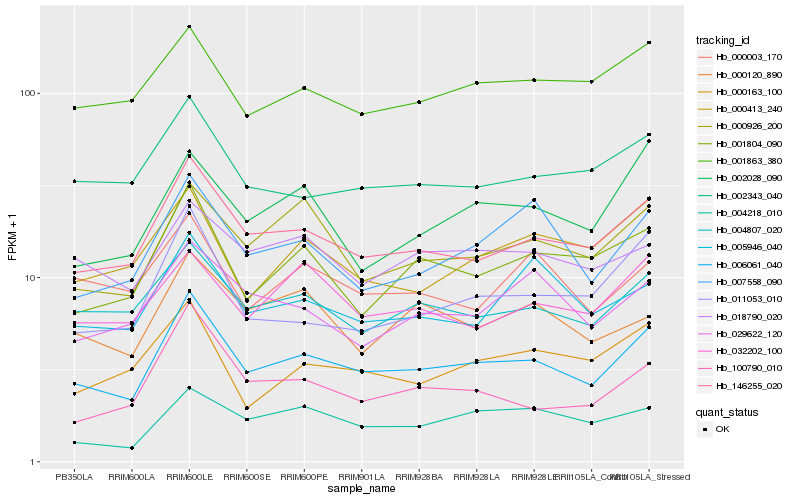
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006061_040 |
0.0 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 2 |
Hb_146255_020 |
0.0674092387 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 3 |
Hb_004807_020 |
0.0974539665 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 4 |
Hb_001804_090 |
0.1023591918 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |
| 5 |
Hb_000926_200 |
0.1054326004 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_011053_010 |
0.1074323082 |
- |
- |
PREDICTED: uncharacterized protein LOC105631146 [Jatropha curcas] |
| 7 |
Hb_002343_040 |
0.1075489786 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001863_380 |
0.1094607188 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 9 |
Hb_007558_090 |
0.1108634942 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_100790_010 |
0.11243367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005946_040 |
0.1170969861 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 12 |
Hb_018790_020 |
0.1181502926 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_002028_090 |
0.118192873 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 14 |
Hb_004218_010 |
0.1186183221 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000413_240 |
0.1187573024 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_032202_100 |
0.1188078891 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 17 |
Hb_029622_120 |
0.119041236 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 18 |
Hb_000163_100 |
0.1191929143 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 19 |
Hb_000120_890 |
0.1218707298 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 20 |
Hb_000003_170 |
0.1224630388 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |