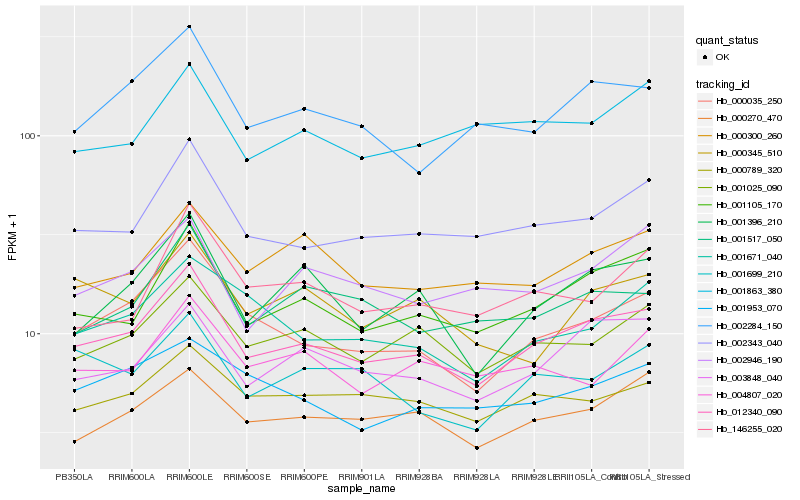
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012340_090 |
0.0 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 2 |
Hb_001105_170 |
0.0663632736 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002343_040 |
0.0704000379 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000035_250 |
0.0735485819 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 5 |
Hb_001025_090 |
0.07687475 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 6 |
Hb_003848_040 |
0.0786621562 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_002284_150 |
0.0864613845 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 8 |
Hb_001396_210 |
0.0872743931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000270_470 |
0.0874740434 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: pentatricopeptide repeat-containing protein At1g74900, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000789_320 |
0.0876604017 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001517_050 |
0.0886548397 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000300_260 |
0.0907704503 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 13 |
Hb_001953_070 |
0.097201676 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001671_040 |
0.0974580131 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 15 |
Hb_002946_190 |
0.0977312468 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004807_020 |
0.0978767474 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 17 |
Hb_001699_210 |
0.0984305637 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001863_380 |
0.0985405484 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 19 |
Hb_146255_020 |
0.1006689614 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 20 |
Hb_000345_510 |
0.1012940469 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |