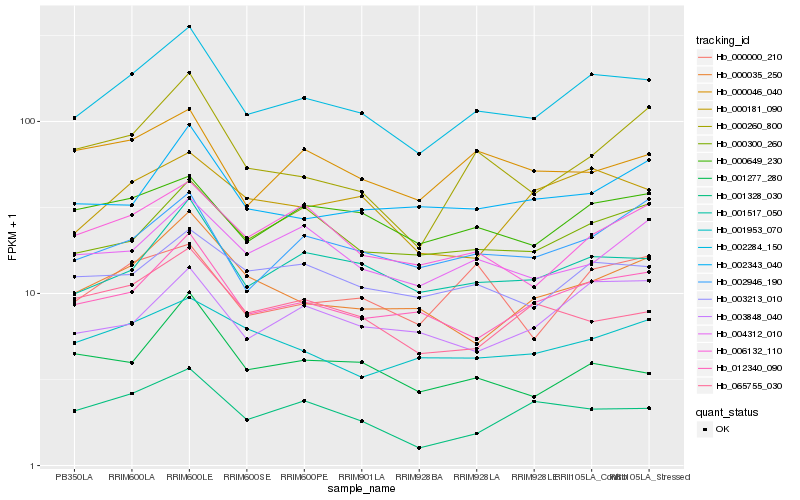
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 2 |
Hb_001517_050 |
0.0794311466 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012340_090 |
0.0864613845 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 4 |
Hb_003213_010 |
0.0979935059 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 5 |
Hb_000300_260 |
0.1016399674 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 6 |
Hb_000649_230 |
0.1018827631 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 7 |
Hb_002946_190 |
0.1074093818 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000035_250 |
0.1079798159 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 9 |
Hb_006132_110 |
0.1086559455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002343_040 |
0.1089807709 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000181_090 |
0.1093487079 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 12 |
Hb_001953_070 |
0.1101206118 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004312_010 |
0.1112483867 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000046_040 |
0.1121290586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003848_040 |
0.112249186 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 16 |
Hb_001328_030 |
0.1132397828 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 17 |
Hb_065755_030 |
0.114034594 |
- |
- |
- |
| 18 |
Hb_000260_800 |
0.114153948 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 19 |
Hb_000000_210 |
0.1143254756 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001277_280 |
0.1144578312 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |