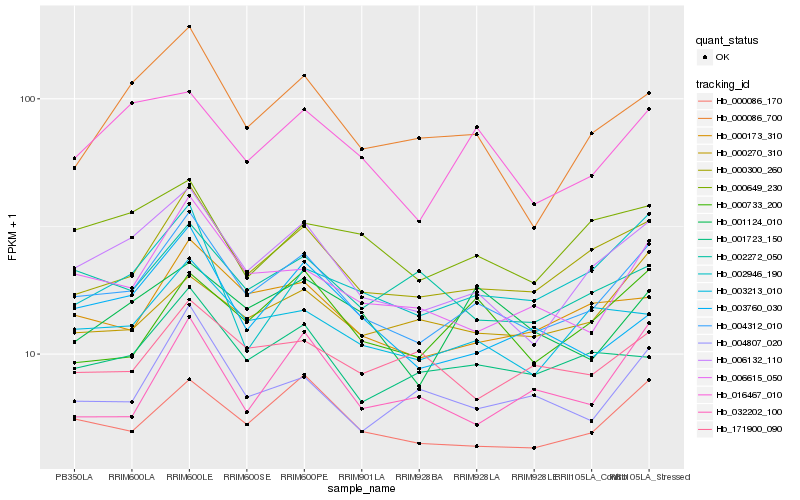
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004312_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 2 |
Hb_006132_110 |
0.0558097265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000300_260 |
0.0574924989 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 4 |
Hb_000086_170 |
0.0685024248 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 5 |
Hb_000173_310 |
0.0729457788 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006615_050 |
0.0740075146 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 7 |
Hb_003213_010 |
0.0766588096 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 8 |
Hb_004807_020 |
0.083684666 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 9 |
Hb_001723_150 |
0.0848110568 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_016467_010 |
0.0852837459 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 11 |
Hb_000270_310 |
0.0909477875 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 12 |
Hb_000649_230 |
0.0915293366 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 13 |
Hb_000733_200 |
0.0919171418 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 14 |
Hb_003760_030 |
0.0926189017 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 15 |
Hb_001124_010 |
0.0950301447 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002272_050 |
0.0950336472 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 17 |
Hb_002946_190 |
0.0952402311 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000086_700 |
0.0952909049 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 19 |
Hb_032202_100 |
0.0965642126 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 20 |
Hb_171900_090 |
0.0981426023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |