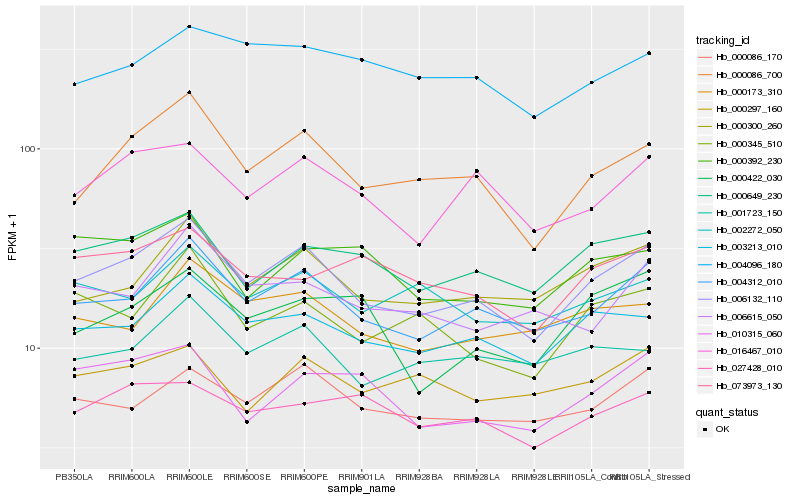
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006132_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004312_010 |
0.0558097265 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000086_700 |
0.0692723131 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 4 |
Hb_003213_010 |
0.0781511815 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 5 |
Hb_000300_260 |
0.0784589118 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 6 |
Hb_000649_230 |
0.0850250429 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 7 |
Hb_016467_010 |
0.0851808998 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 8 |
Hb_000086_170 |
0.0876514257 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 9 |
Hb_000345_510 |
0.094608998 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 10 |
Hb_000173_310 |
0.0955557015 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001723_150 |
0.0973368823 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_010315_060 |
0.0988786008 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 13 |
Hb_002272_050 |
0.1008612802 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 14 |
Hb_006615_050 |
0.1022131858 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 15 |
Hb_027428_010 |
0.103782637 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_004096_180 |
0.1047861192 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 17 |
Hb_000297_160 |
0.1049373596 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 18 |
Hb_000392_230 |
0.1055688545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000422_030 |
0.1060059039 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 20 |
Hb_073973_130 |
0.1060836324 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |