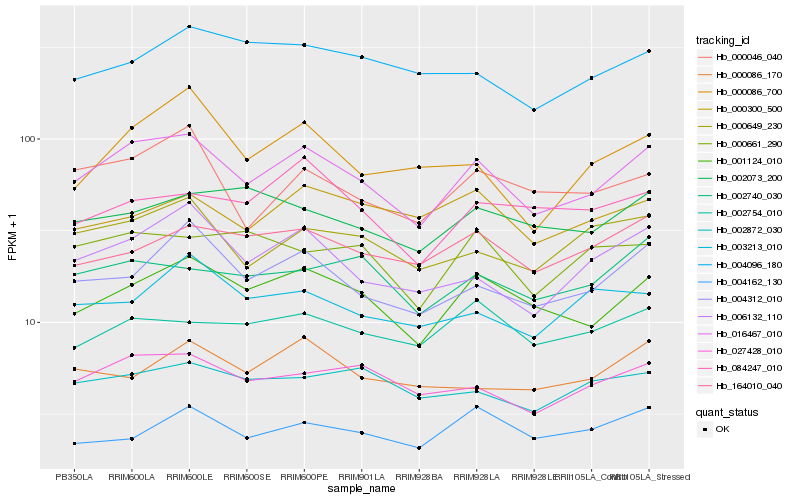
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016467_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 2 |
Hb_001124_010 |
0.0698230614 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 3 |
Hb_006132_110 |
0.0851808998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004312_010 |
0.0852837459 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 5 |
Hb_027428_010 |
0.0861011668 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000649_230 |
0.0910070493 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 7 |
Hb_000046_040 |
0.0955706181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_164010_040 |
0.1010270561 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_002740_030 |
0.1015340579 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002754_010 |
0.1034268255 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 11 |
Hb_002872_030 |
0.1066867157 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 12 |
Hb_000661_290 |
0.1076519299 |
- |
- |
PREDICTED: NAD(H) kinase 1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_002073_200 |
0.1078859887 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 14 |
Hb_000086_170 |
0.1081052389 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 15 |
Hb_000086_700 |
0.1082527792 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 16 |
Hb_003213_010 |
0.1084758953 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 17 |
Hb_000300_500 |
0.1085647088 |
- |
- |
PREDICTED: uncharacterized protein LOC105632604 [Jatropha curcas] |
| 18 |
Hb_084247_010 |
0.1089045419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004096_180 |
0.1091026772 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 20 |
Hb_004162_130 |
0.1091924227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |