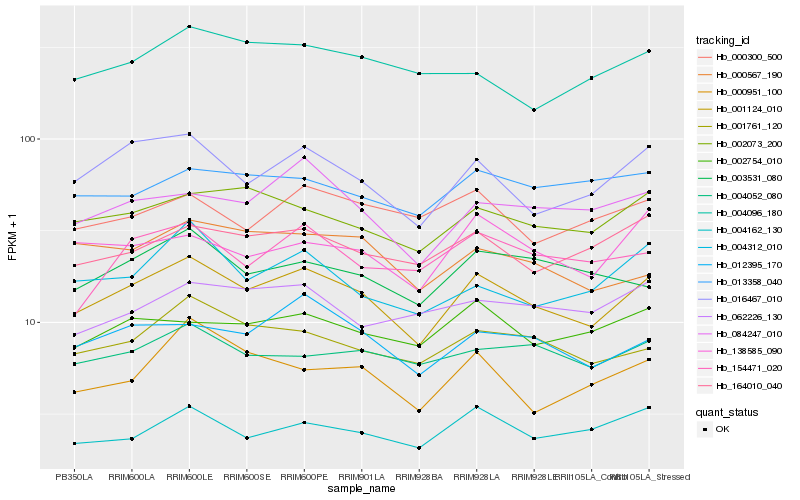
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 2 |
Hb_016467_010 |
0.0698230614 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 3 |
Hb_012395_170 |
0.0811992266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001761_120 |
0.0835986924 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 5 |
Hb_002073_200 |
0.0843405738 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 6 |
Hb_084247_010 |
0.0881842678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_164010_040 |
0.0897488084 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_004162_130 |
0.0903182783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002754_010 |
0.0924321016 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 10 |
Hb_138585_090 |
0.093651466 |
- |
- |
Long chain base2 isoform 2 [Theobroma cacao] |
| 11 |
Hb_003531_080 |
0.0941211832 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004312_010 |
0.0950301447 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004052_080 |
0.095994298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_154471_020 |
0.0970088208 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |
| 15 |
Hb_000567_190 |
0.0977919223 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_013358_040 |
0.0981133233 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 17 |
Hb_000951_100 |
0.1002326081 |
- |
- |
PREDICTED: uncharacterized protein LOC103436565 isoform X1 [Malus domestica] |
| 18 |
Hb_000300_500 |
0.1002951447 |
- |
- |
PREDICTED: uncharacterized protein LOC105632604 [Jatropha curcas] |
| 19 |
Hb_062226_130 |
0.1012801751 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 20 |
Hb_004096_180 |
0.1021390936 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |