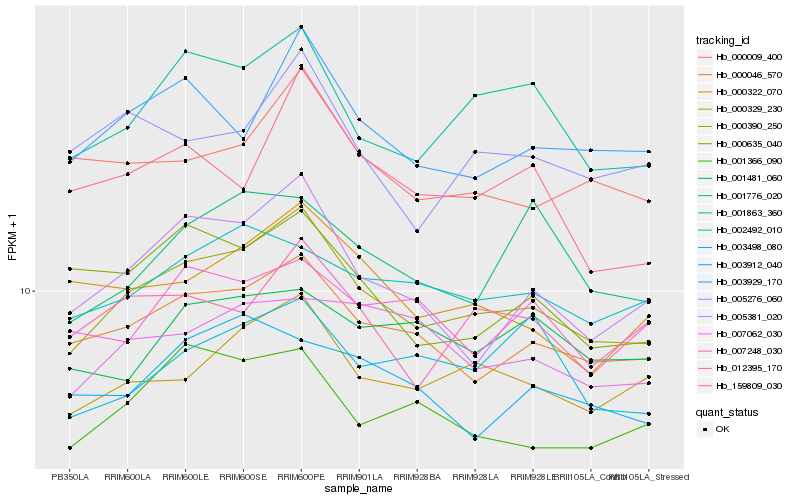
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_250 |
0.0 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 2 |
Hb_000046_570 |
0.05902177 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 3 |
Hb_000329_230 |
0.0673462745 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 4 |
Hb_001863_360 |
0.0674355642 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 5 |
Hb_005381_020 |
0.0718993833 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 6 |
Hb_003929_170 |
0.075806169 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 7 |
Hb_001366_090 |
0.079435086 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 8 |
Hb_002492_010 |
0.0799546817 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 9 |
Hb_000009_400 |
0.081070236 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 10 |
Hb_000322_070 |
0.0811092008 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_012395_170 |
0.0829360501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007062_030 |
0.0860628759 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 13 |
Hb_000635_040 |
0.0860921926 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 14 |
Hb_005276_060 |
0.0861997065 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_159809_030 |
0.0890155463 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_003912_040 |
0.0896220554 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 17 |
Hb_003498_080 |
0.0902600423 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_001776_020 |
0.0903635132 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 19 |
Hb_007248_030 |
0.0910635194 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 20 |
Hb_001481_060 |
0.0914973987 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |