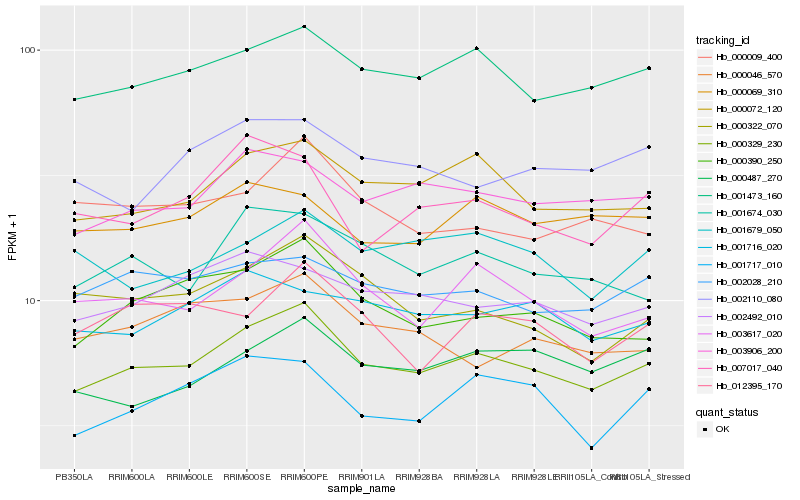
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 2 |
Hb_001473_160 |
0.0564048742 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 3 |
Hb_000072_120 |
0.0606114607 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_003617_020 |
0.0655905354 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 5 |
Hb_000390_250 |
0.0673462745 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 6 |
Hb_003906_200 |
0.0682857649 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 7 |
Hb_001674_030 |
0.0717589779 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 8 |
Hb_002492_010 |
0.075063053 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 9 |
Hb_002028_210 |
0.0775513403 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |
| 10 |
Hb_001717_010 |
0.0803557594 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 11 |
Hb_001716_020 |
0.0815152838 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 12 |
Hb_000487_270 |
0.0818801796 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 13 |
Hb_000322_070 |
0.0819555967 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_000009_400 |
0.0837807847 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 15 |
Hb_007017_040 |
0.0839775697 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 16 |
Hb_012395_170 |
0.0841591434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002110_080 |
0.0846335598 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 18 |
Hb_000046_570 |
0.084925356 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 19 |
Hb_001679_050 |
0.0858078949 |
- |
- |
PREDICTED: exocyst complex component EXO84B [Jatropha curcas] |
| 20 |
Hb_000069_310 |
0.0858907749 |
- |
- |
ubiquitin ligase [Cucumis melo subsp. melo] |