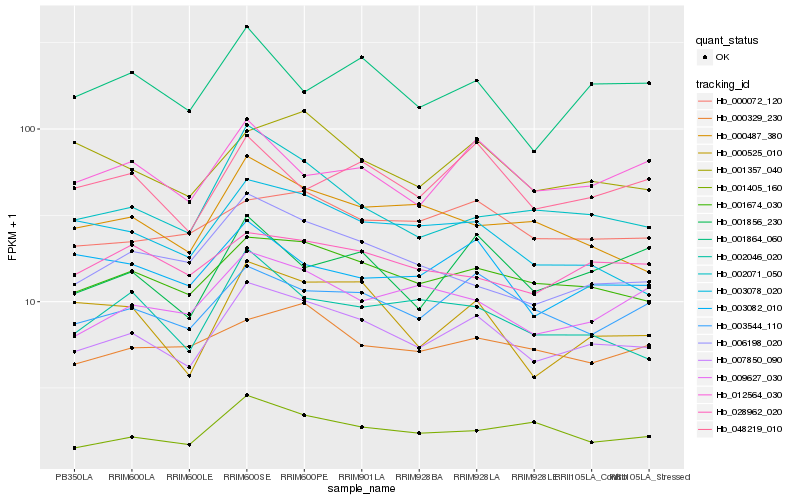
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007850_090 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001674_030 |
0.069259299 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 3 |
Hb_003544_110 |
0.0897588575 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000525_010 |
0.1008437099 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_000072_120 |
0.1010292255 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 6 |
Hb_003078_020 |
0.1011926969 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 7 |
Hb_009627_030 |
0.1027312401 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 8 |
Hb_002071_050 |
0.1027353929 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 9 |
Hb_001357_040 |
0.1028669434 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 10 |
Hb_012564_030 |
0.1045960759 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 11 |
Hb_002046_020 |
0.1056574224 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 12 |
Hb_048219_010 |
0.1060369636 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 13 |
Hb_000487_380 |
0.1063694988 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006198_020 |
0.1070093176 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 15 |
Hb_001405_160 |
0.107599015 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 16 |
Hb_003082_010 |
0.1091019975 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 17 |
Hb_000329_230 |
0.1091968669 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 18 |
Hb_001864_060 |
0.1092676142 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 19 |
Hb_028962_020 |
0.1094677116 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 20 |
Hb_001856_230 |
0.1096638962 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |