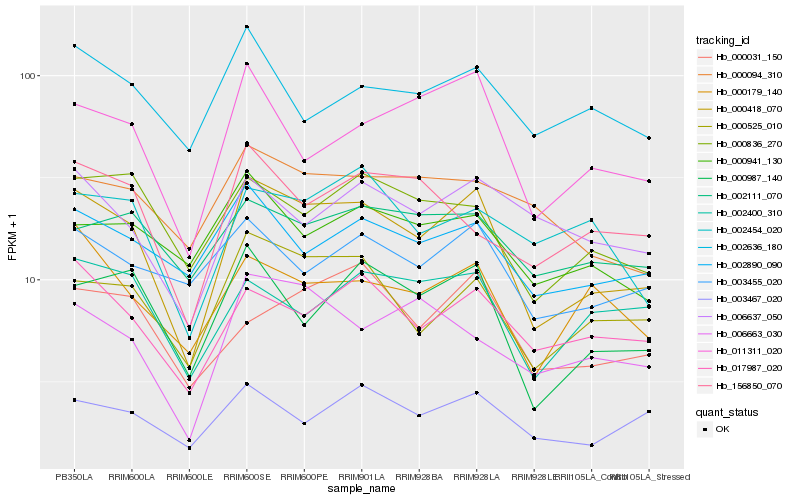
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000418_070 |
0.0 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 2 |
Hb_000525_010 |
0.1147781541 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_000987_140 |
0.1193385859 |
- |
- |
PREDICTED: uncharacterized protein LOC105648083 isoform X1 [Jatropha curcas] |
| 4 |
Hb_017987_020 |
0.1242405108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011311_020 |
0.12621623 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000836_270 |
0.1361037787 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 7 |
Hb_000179_140 |
0.1375265083 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000031_150 |
0.1379734328 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 9 |
Hb_002890_090 |
0.1385458743 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 10 |
Hb_003467_020 |
0.1404758443 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 13-like [Jatropha curcas] |
| 11 |
Hb_003455_020 |
0.1431921082 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 12 |
Hb_002400_310 |
0.1437334725 |
- |
- |
hypothetical protein JCGZ_07138 [Jatropha curcas] |
| 13 |
Hb_006637_050 |
0.1443408754 |
- |
- |
PREDICTED: ethylene-insensitive protein 2 [Jatropha curcas] |
| 14 |
Hb_006663_030 |
0.1457124887 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_156850_070 |
0.1482651351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002454_020 |
0.1496660464 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 9 [Vitis vinifera] |
| 17 |
Hb_000941_130 |
0.1501146518 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 18 |
Hb_002636_180 |
0.1506909114 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 19 |
Hb_000094_310 |
0.1520515092 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 20 |
Hb_002111_070 |
0.1523879498 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |