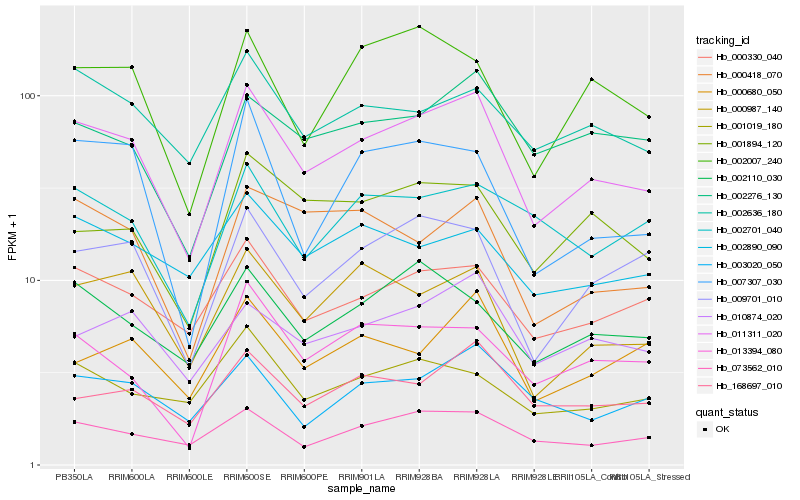
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011311_020 |
0.0 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000330_040 |
0.1052447921 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 3 |
Hb_009701_010 |
0.1089304739 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 4 |
Hb_073562_010 |
0.109265183 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 5 |
Hb_000987_140 |
0.1122549665 |
- |
- |
PREDICTED: uncharacterized protein LOC105648083 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010874_020 |
0.1145172008 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |
| 7 |
Hb_168697_010 |
0.1172979981 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 8 |
Hb_002636_180 |
0.1205858306 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 9 |
Hb_002110_030 |
0.1213716032 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_002276_130 |
0.1230749072 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 11 |
Hb_003020_050 |
0.1239934318 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001894_120 |
0.124088839 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 13 |
Hb_002007_240 |
0.1247505871 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 14 |
Hb_000418_070 |
0.12621623 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 15 |
Hb_007307_030 |
0.1267937977 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 16 |
Hb_013394_080 |
0.1304085807 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1D isoform X2 [Jatropha curcas] |
| 17 |
Hb_001019_180 |
0.131238692 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 18 |
Hb_002701_040 |
0.131586695 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 19 |
Hb_000680_050 |
0.1319024437 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 20 |
Hb_002890_090 |
0.1332513564 |
- |
- |
Auxin response factor 1 [Glycine soja] |