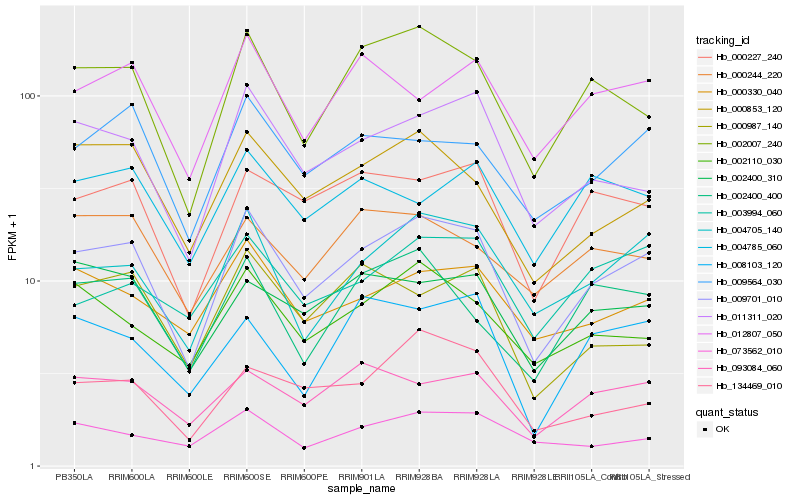
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009701_010 |
0.0 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 2 |
Hb_002007_240 |
0.0966124951 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 3 |
Hb_004705_140 |
0.1007011542 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 4 |
Hb_011311_020 |
0.1089304739 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000227_240 |
0.111332949 |
- |
- |
hypothetical protein JCGZ_02139 [Jatropha curcas] |
| 6 |
Hb_000853_120 |
0.1139636901 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 7 |
Hb_009564_030 |
0.1145615061 |
transcription factor |
TF Family: bHLH |
Transcription factor AtMYC2, putative [Ricinus communis] |
| 8 |
Hb_000330_040 |
0.1218801186 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 9 |
Hb_093084_060 |
0.1218961351 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |
| 10 |
Hb_002110_030 |
0.1246849771 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 11 |
Hb_002400_310 |
0.1247663924 |
- |
- |
hypothetical protein JCGZ_07138 [Jatropha curcas] |
| 12 |
Hb_012807_050 |
0.1273714344 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 13 |
Hb_134469_010 |
0.1294668006 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002400_400 |
0.1301152562 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 15 |
Hb_003994_060 |
0.1358992793 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 16 |
Hb_073562_010 |
0.1359174336 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 17 |
Hb_000244_220 |
0.1371203225 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_008103_120 |
0.1374108858 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 19 |
Hb_000987_140 |
0.1385214839 |
- |
- |
PREDICTED: uncharacterized protein LOC105648083 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004785_060 |
0.1399097296 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |