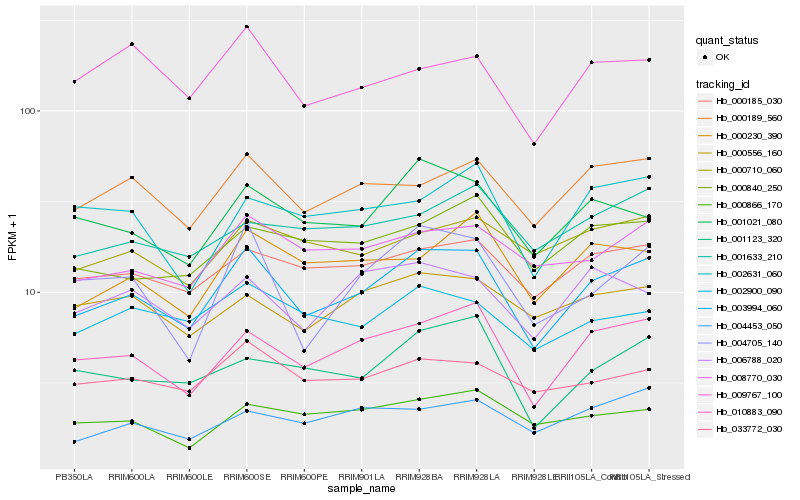
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 2 |
Hb_010883_090 |
0.0750315134 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000189_560 |
0.0945734158 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 4 |
Hb_000185_030 |
0.1001845382 |
- |
- |
PREDICTED: arginine-glutamic acid dipeptide repeats protein-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001021_080 |
0.1009015733 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 6 |
Hb_008770_030 |
0.101464039 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290-like [Jatropha curcas] |
| 7 |
Hb_004705_140 |
0.1059013065 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 8 |
Hb_000230_390 |
0.1064907287 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 9 |
Hb_033772_030 |
0.1075691653 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 10 |
Hb_001123_320 |
0.109574314 |
- |
- |
PREDICTED: uncharacterized protein LOC105647314 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000840_250 |
0.111453936 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 12 |
Hb_006788_020 |
0.1116815369 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 13 |
Hb_002900_090 |
0.1131398326 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 14 |
Hb_004453_050 |
0.1138302837 |
- |
- |
PREDICTED: uncharacterized protein LOC105637675 [Jatropha curcas] |
| 15 |
Hb_000710_060 |
0.1142440538 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105645554 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000556_160 |
0.1148103348 |
- |
- |
PREDICTED: ankyrin repeat and zinc finger domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_000866_170 |
0.114965419 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_009767_100 |
0.1157013754 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 19 |
Hb_001633_210 |
0.1165795128 |
- |
- |
PREDICTED: RING finger protein 10 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002631_060 |
0.1169757855 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |