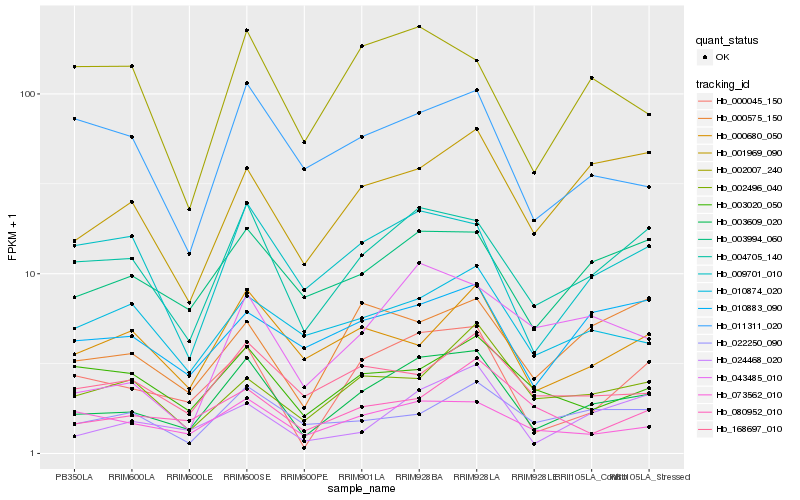
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003609_020 |
0.0 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 2 |
Hb_004705_140 |
0.1217654144 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 3 |
Hb_000045_150 |
0.1345923695 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 4 |
Hb_003994_060 |
0.1551205648 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 5 |
Hb_073562_010 |
0.16379084 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 6 |
Hb_024468_020 |
0.1668320582 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 7 |
Hb_001969_090 |
0.1675246546 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 8 |
Hb_009701_010 |
0.1682006159 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 9 |
Hb_168697_010 |
0.172279976 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 10 |
Hb_043485_010 |
0.1752716484 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_010883_090 |
0.1784001037 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000680_050 |
0.1795181512 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 13 |
Hb_000575_150 |
0.181199829 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_011311_020 |
0.1821257927 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002496_040 |
0.183291626 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 16 |
Hb_010874_020 |
0.1844060425 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |
| 17 |
Hb_002007_240 |
0.1844941892 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 18 |
Hb_003020_050 |
0.185181229 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_080952_010 |
0.1864715165 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |
| 20 |
Hb_022250_090 |
0.1866916833 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |