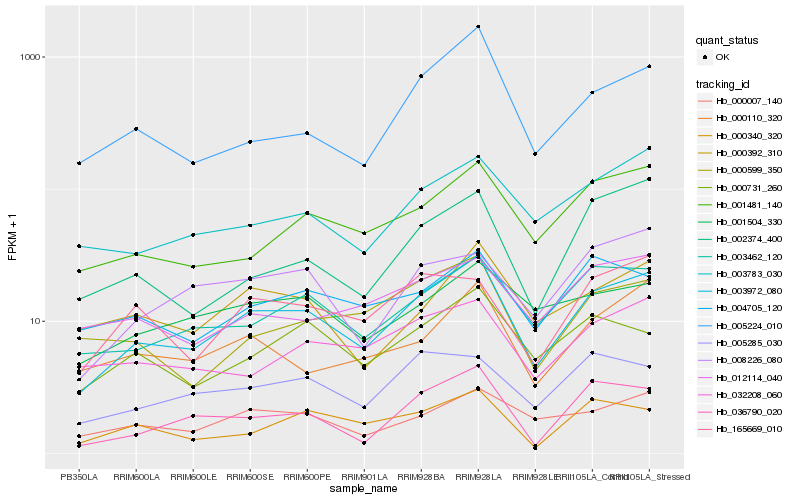
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003972_080 |
0.0 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 2 |
Hb_003462_120 |
0.1191619725 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 3 |
Hb_001481_140 |
0.1279716801 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 4 |
Hb_003783_030 |
0.1305472763 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 5 |
Hb_000007_140 |
0.1323995633 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 6 |
Hb_005224_010 |
0.1356201263 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 7 |
Hb_036790_020 |
0.1371201456 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 8 |
Hb_165669_010 |
0.1424916774 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000110_320 |
0.1425951338 |
- |
- |
PREDICTED: ceramide-1-phosphate transfer protein [Jatropha curcas] |
| 10 |
Hb_008226_080 |
0.1426461834 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 11 |
Hb_000392_310 |
0.1428320986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000731_260 |
0.1434064451 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 13 |
Hb_032208_060 |
0.143795036 |
- |
- |
PREDICTED: constitutive photomorphogenesis protein 10 isoform X2 [Populus euphratica] |
| 14 |
Hb_002374_400 |
0.1438073118 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_005285_030 |
0.1447274606 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 16 |
Hb_001504_330 |
0.1457858874 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 17 |
Hb_004705_120 |
0.1466695454 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_012114_040 |
0.1480247022 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000599_350 |
0.1482293895 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_000340_320 |
0.1494805299 |
- |
- |
PREDICTED: uncharacterized protein LOC105650052 [Jatropha curcas] |