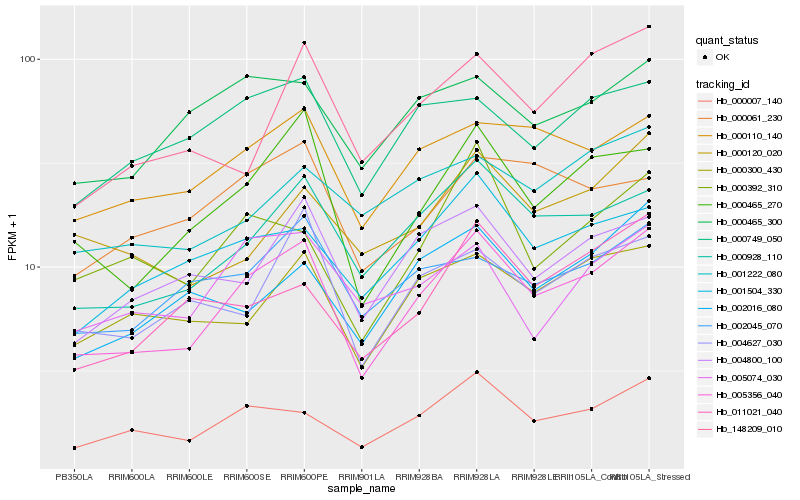
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005356_040 |
0.0 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000465_270 |
0.0937877873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000928_110 |
0.0950740928 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000007_140 |
0.0970409052 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 5 |
Hb_000110_140 |
0.1127148614 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 6 |
Hb_004800_100 |
0.117932048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001504_330 |
0.1184439541 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 8 |
Hb_000300_430 |
0.1197337531 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 9 |
Hb_004627_030 |
0.1245278211 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 10 |
Hb_000749_050 |
0.1255878315 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 11 |
Hb_002045_070 |
0.1275769238 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 12 |
Hb_000465_300 |
0.1293109372 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 13 |
Hb_002016_080 |
0.1295292914 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000392_310 |
0.1311314763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011021_040 |
0.1316275045 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 16 |
Hb_148209_010 |
0.132640915 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 17 |
Hb_000120_020 |
0.1345135812 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 18 |
Hb_005074_030 |
0.1347491891 |
- |
- |
- |
| 19 |
Hb_001222_080 |
0.1360394816 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000061_230 |
0.1366254842 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |