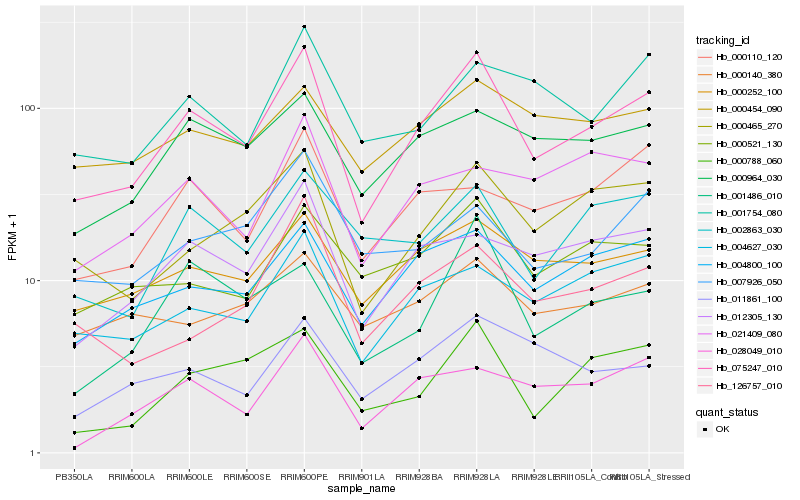
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075247_010 |
0.0 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 2 |
Hb_004800_100 |
0.1234189123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000465_270 |
0.1254398438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004627_030 |
0.1309065641 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 5 |
Hb_012305_130 |
0.1314573983 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 6 |
Hb_000252_100 |
0.1335435131 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 7 |
Hb_000788_060 |
0.1393522014 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 8 |
Hb_000964_030 |
0.1418423129 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 9 |
Hb_000521_130 |
0.1427696154 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002863_030 |
0.1440319192 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 11 |
Hb_001486_010 |
0.1474608192 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 12 |
Hb_001754_080 |
0.1478913335 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 13 |
Hb_126757_010 |
0.150455074 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_011861_100 |
0.1507346053 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_028049_010 |
0.1512283132 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000110_120 |
0.1516584501 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 17 |
Hb_000454_090 |
0.1529581353 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 18 |
Hb_007926_050 |
0.1536856206 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 19 |
Hb_000140_380 |
0.1546569825 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 20 |
Hb_021409_080 |
0.1558342155 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |