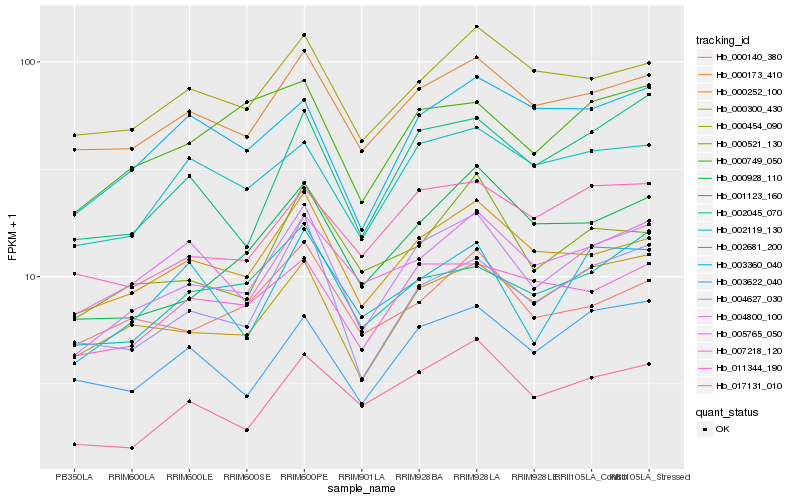
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000252_100 |
0.0775322698 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 3 |
Hb_004627_030 |
0.0787461282 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 4 |
Hb_000300_430 |
0.0792393829 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 5 |
Hb_000173_410 |
0.0796265416 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 6 |
Hb_001123_160 |
0.0897273067 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 7 |
Hb_002045_070 |
0.0933456919 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 8 |
Hb_017131_010 |
0.0943847157 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 9 |
Hb_000521_130 |
0.0946357217 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000928_110 |
0.0949390132 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007218_120 |
0.0959747694 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000454_090 |
0.0962335871 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 13 |
Hb_002119_130 |
0.098070475 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 14 |
Hb_000749_050 |
0.0985457775 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 15 |
Hb_011344_190 |
0.0988680928 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 16 |
Hb_003622_040 |
0.0990047444 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 17 |
Hb_000140_380 |
0.0997817254 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 18 |
Hb_005765_050 |
0.1030914737 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4-like [Populus euphratica] |
| 19 |
Hb_002681_200 |
0.1035634011 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 20 |
Hb_003360_040 |
0.1044509166 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |