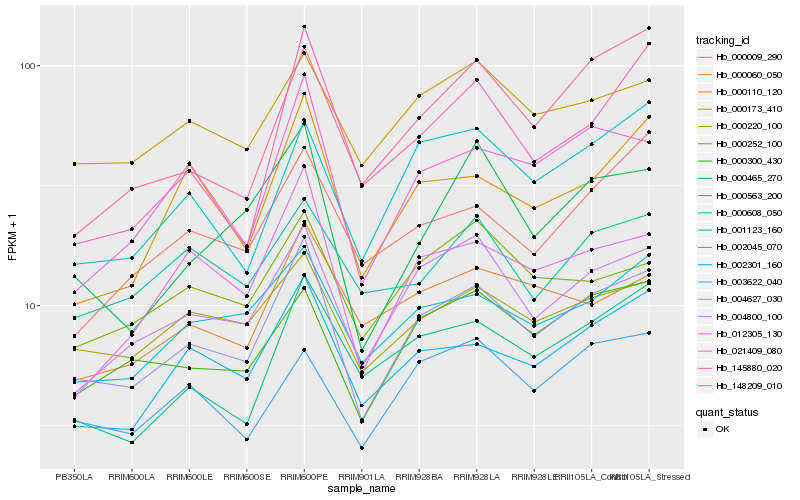
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004627_030 |
0.0 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 2 |
Hb_004800_100 |
0.0787461282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002301_160 |
0.0804647012 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 4 |
Hb_021409_080 |
0.088663517 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 5 |
Hb_002045_070 |
0.0902116819 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 6 |
Hb_000300_430 |
0.0903680908 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 7 |
Hb_012305_130 |
0.09444785 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 8 |
Hb_000220_100 |
0.0956162461 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 [Jatropha curcas] |
| 9 |
Hb_001123_160 |
0.097057852 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_000110_120 |
0.0984720012 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 11 |
Hb_000173_410 |
0.0994069666 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 12 |
Hb_000465_270 |
0.1018028442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000563_200 |
0.1024022634 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |
| 14 |
Hb_003622_040 |
0.1058488039 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 15 |
Hb_000252_100 |
0.1059029827 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 16 |
Hb_148209_010 |
0.1083854246 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 17 |
Hb_000060_050 |
0.11101656 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 18 |
Hb_000009_290 |
0.111114201 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 19 |
Hb_145880_020 |
0.1148934048 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 20 |
Hb_000608_050 |
0.1154783397 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |