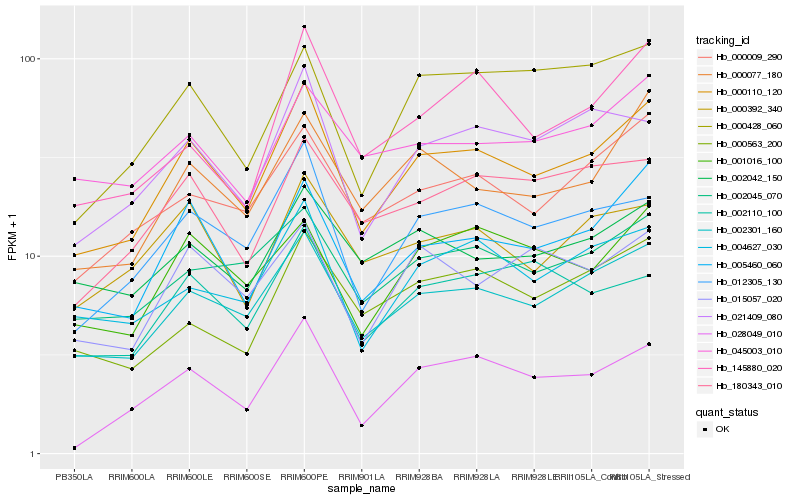
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_120 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 2 |
Hb_002301_160 |
0.0619810125 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 3 |
Hb_005460_060 |
0.0783672016 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 4 |
Hb_012305_130 |
0.0835674353 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 5 |
Hb_000009_290 |
0.0981423117 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 6 |
Hb_004627_030 |
0.0984720012 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 7 |
Hb_000077_180 |
0.0984986448 |
- |
- |
- |
| 8 |
Hb_021409_080 |
0.1000413732 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 9 |
Hb_028049_010 |
0.1038496645 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002045_070 |
0.1054262141 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 11 |
Hb_001016_100 |
0.1091818069 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 12 |
Hb_000392_340 |
0.1095437099 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 13 |
Hb_180343_010 |
0.1102914602 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 14 |
Hb_015057_020 |
0.1123298439 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 15 |
Hb_145880_020 |
0.1128418887 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 16 |
Hb_002042_150 |
0.1135070464 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 17 |
Hb_000428_060 |
0.1137501067 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_002110_100 |
0.1150817283 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_045003_010 |
0.1159319721 |
- |
- |
Stomatin-1, putative [Ricinus communis] |
| 20 |
Hb_000563_200 |
0.1161455533 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |