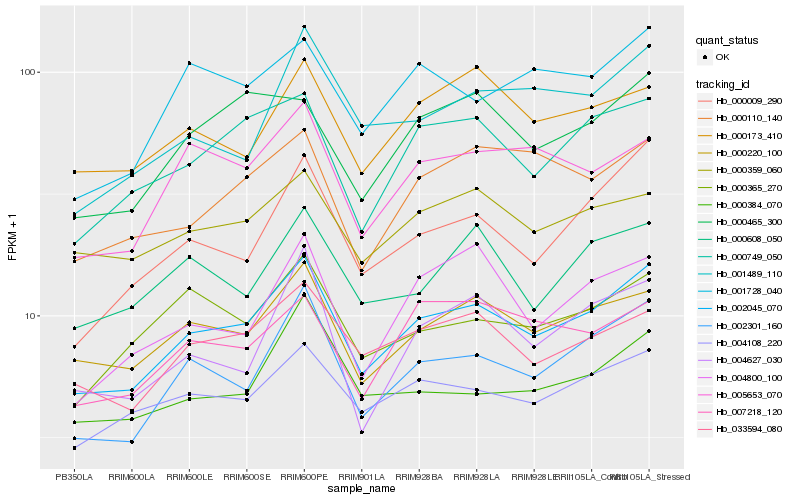
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_070 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 2 |
Hb_002301_160 |
0.0611028112 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 3 |
Hb_000220_100 |
0.0682623945 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 [Jatropha curcas] |
| 4 |
Hb_004108_220 |
0.073334301 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_033594_080 |
0.0743465517 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 6 |
Hb_000749_050 |
0.0756615071 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 7 |
Hb_000009_290 |
0.0787615437 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 8 |
Hb_005653_070 |
0.0847555684 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 9 |
Hb_000365_270 |
0.0866732308 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 10 |
Hb_000359_060 |
0.0872209549 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 11 |
Hb_000465_300 |
0.0881251543 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 12 |
Hb_000173_410 |
0.0889529486 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 13 |
Hb_007218_120 |
0.0892433321 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001489_110 |
0.0892649031 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 15 |
Hb_000384_070 |
0.0895898059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000110_140 |
0.0897356361 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 17 |
Hb_001728_040 |
0.0901586695 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 18 |
Hb_004627_030 |
0.0902116819 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 19 |
Hb_000608_050 |
0.0908639022 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 20 |
Hb_004800_100 |
0.0933456919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |