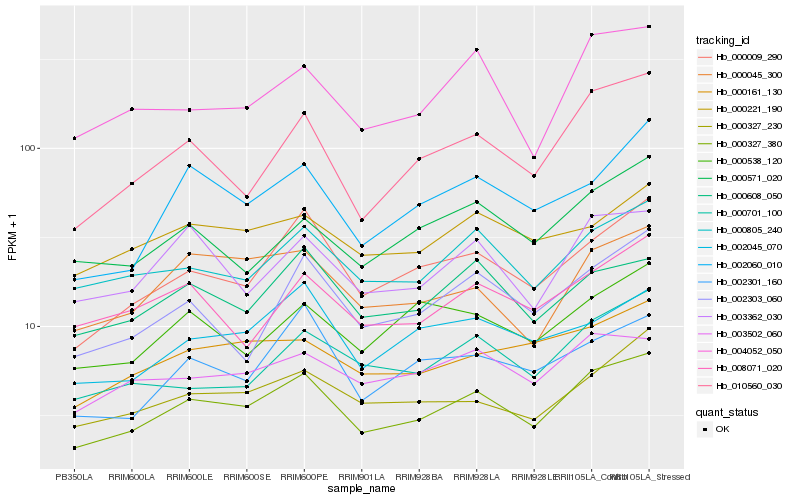
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_380 |
0.0 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 2 |
Hb_000009_290 |
0.075825443 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 3 |
Hb_000805_240 |
0.0810675154 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 4 |
Hb_010560_030 |
0.0852354898 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 5 |
Hb_000327_230 |
0.0898478493 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003362_030 |
0.0917448396 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 7 |
Hb_000045_300 |
0.092379381 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 8 |
Hb_004052_050 |
0.092957467 |
- |
- |
PREDICTED: uncharacterized protein LOC105640640 [Jatropha curcas] |
| 9 |
Hb_000608_050 |
0.0937436298 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 10 |
Hb_002060_010 |
0.0970951676 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002303_060 |
0.0980878107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002045_070 |
0.0989798025 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 13 |
Hb_003502_060 |
0.0997204681 |
- |
- |
PREDICTED: alpha-1,3/1,6-mannosyltransferase ALG2 [Jatropha curcas] |
| 14 |
Hb_002301_160 |
0.1001848167 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 15 |
Hb_000701_100 |
0.1008315791 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 16 |
Hb_000161_130 |
0.1010096973 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000571_020 |
0.1037524709 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008071_020 |
0.1042805322 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC1 [Jatropha curcas] |
| 19 |
Hb_000538_120 |
0.1044511722 |
transcription factor |
TF Family: SET |
PREDICTED: protein SET DOMAIN GROUP 40 [Jatropha curcas] |
| 20 |
Hb_000221_190 |
0.1050003485 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |