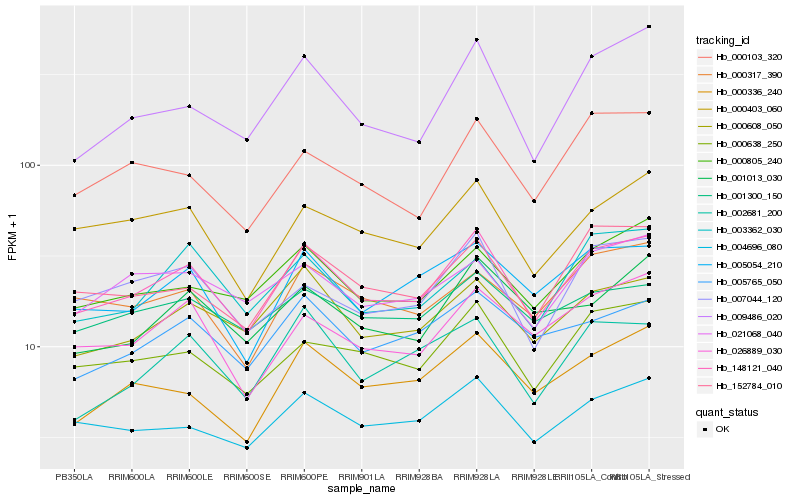
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148121_040 |
0.0 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000608_050 |
0.0711856533 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 3 |
Hb_000638_250 |
0.0742628086 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_004696_080 |
0.0742814302 |
- |
- |
PREDICTED: uncharacterized protein At1g01500-like [Populus euphratica] |
| 5 |
Hb_152784_010 |
0.0747208434 |
- |
- |
hypothetical protein PRUPE_ppa010913mg [Prunus persica] |
| 6 |
Hb_005054_210 |
0.0771823654 |
- |
- |
hypothetical protein PRUPE_ppa026456mg [Prunus persica] |
| 7 |
Hb_000403_060 |
0.0807859458 |
- |
- |
PREDICTED: probable ATP synthase 24 kDa subunit, mitochondrial [Vitis vinifera] |
| 8 |
Hb_005765_050 |
0.0817444855 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4-like [Populus euphratica] |
| 9 |
Hb_000805_240 |
0.0820394865 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 10 |
Hb_026889_030 |
0.0829267349 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_007044_120 |
0.0847349296 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 12 |
Hb_002681_200 |
0.0854870863 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 13 |
Hb_000336_240 |
0.0868582607 |
transcription factor |
TF Family: mTERF |
LOC100285792 [Zea mays] |
| 14 |
Hb_009486_020 |
0.0880549186 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 15 |
Hb_000317_390 |
0.0898149248 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 16 |
Hb_021068_040 |
0.0901595938 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 17 |
Hb_001013_030 |
0.0906286866 |
- |
- |
GTP-binding protein yptv3, putative [Ricinus communis] |
| 18 |
Hb_003362_030 |
0.0906357186 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 19 |
Hb_000103_320 |
0.0907685302 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 20 |
Hb_001300_150 |
0.0910079876 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |