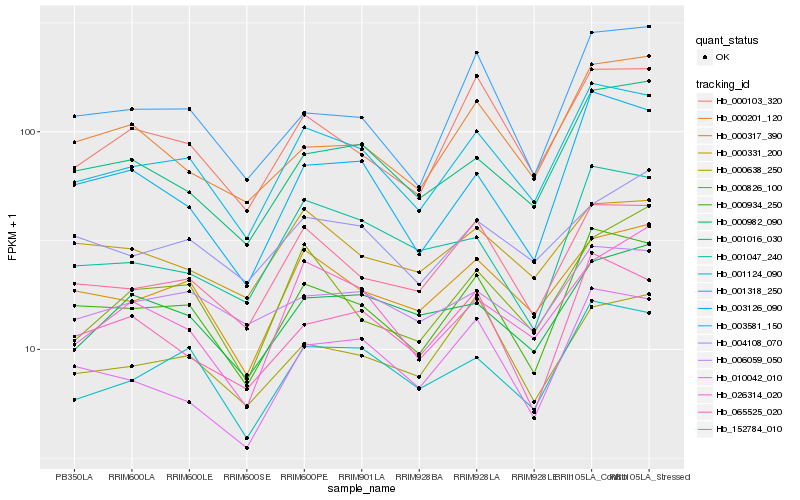
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642666 [Jatropha curcas] |
| 2 |
Hb_000934_250 |
0.0461437559 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 3 |
Hb_001124_090 |
0.0555668259 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 4 |
Hb_152784_010 |
0.0645776611 |
- |
- |
hypothetical protein PRUPE_ppa010913mg [Prunus persica] |
| 5 |
Hb_000317_390 |
0.066955582 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 6 |
Hb_000103_320 |
0.0730256627 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 7 |
Hb_001016_030 |
0.0751180223 |
- |
- |
PREDICTED: uncharacterized protein LOC105636529 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003126_090 |
0.0828782052 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 9 |
Hb_010042_010 |
0.0836115469 |
- |
- |
Urease accessory protein ureD, putative [Ricinus communis] |
| 10 |
Hb_003581_150 |
0.0881750377 |
- |
- |
hypothetical protein CICLE_v10032366mg [Citrus clementina] |
| 11 |
Hb_006059_050 |
0.0882370595 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 12 |
Hb_026314_020 |
0.0885205809 |
- |
- |
hypothetical protein POPTR_0019s10050g [Populus trichocarpa] |
| 13 |
Hb_000638_250 |
0.0885691224 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_001047_240 |
0.0888092921 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 20-like [Jatropha curcas] |
| 15 |
Hb_000982_090 |
0.0888095248 |
- |
- |
PREDICTED: zinc finger HIT domain-containing protein 3 [Jatropha curcas] |
| 16 |
Hb_065525_020 |
0.0889992513 |
- |
- |
PREDICTED: geranylgeranyl transferase type-1 subunit beta [Jatropha curcas] |
| 17 |
Hb_000331_200 |
0.0899372509 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 18 |
Hb_004108_070 |
0.0903572565 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Jatropha curcas] |
| 19 |
Hb_000826_100 |
0.0904468576 |
- |
- |
PREDICTED: biogenesis of lysosome-related organelles complex 1 subunit 1 [Jatropha curcas] |
| 20 |
Hb_000201_120 |
0.0906787322 |
- |
- |
ubiquitin-protein ligase 2a [Hevea brasiliensis] |