| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
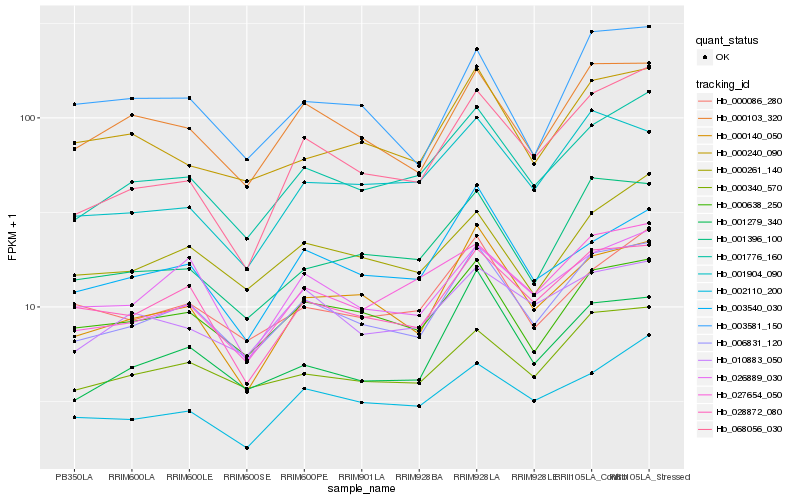
Hb_006831_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642121 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001776_160 |
0.0576894095 |
- |
- |
20S proteasome alpha subunit G1 [Theobroma cacao] |
| 3 |
Hb_000103_320 |
0.0645307667 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 4 |
Hb_000638_250 |
0.0675412696 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_000340_570 |
0.069066668 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 6 |
Hb_028872_080 |
0.0701765703 |
- |
- |
PREDICTED: adenylate kinase 4-like [Jatropha curcas] |
| 7 |
Hb_000140_050 |
0.0707555827 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001396_100 |
0.077776911 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_001279_340 |
0.078743999 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 10 |
Hb_000086_280 |
0.0808229871 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 3-like [Gossypium raimondii] |
| 11 |
Hb_000261_140 |
0.0816183071 |
- |
- |
- |
| 12 |
Hb_026889_030 |
0.08505571 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000240_090 |
0.0861513982 |
- |
- |
Rab5 [Hevea brasiliensis] |
| 14 |
Hb_027654_050 |
0.0863946961 |
- |
- |
PREDICTED: uncharacterized protein LOC105646592 [Jatropha curcas] |
| 15 |
Hb_003540_030 |
0.0865061705 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 16 |
Hb_068056_030 |
0.0869036982 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 17 |
Hb_010883_050 |
0.08770744 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001904_090 |
0.0888559494 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 19 |
Hb_002110_200 |
0.0892145996 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_003581_150 |
0.0896640544 |
- |
- |
hypothetical protein CICLE_v10032366mg [Citrus clementina] |