| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
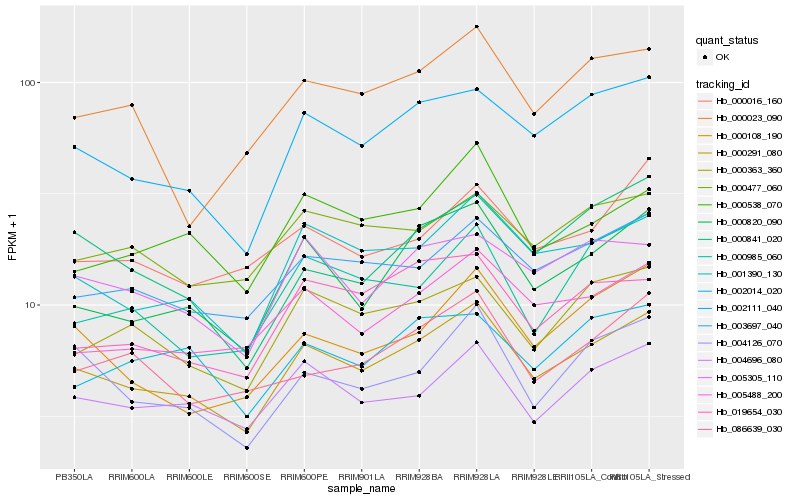
Hb_000291_080 |
0.0 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 2 |
Hb_001390_130 |
0.060965013 |
- |
- |
hypothetical protein POPTR_0009s08590g [Populus trichocarpa] |
| 3 |
Hb_002111_040 |
0.0619482746 |
- |
- |
hypothetical protein JCGZ_16989 [Jatropha curcas] |
| 4 |
Hb_000820_090 |
0.0680805694 |
- |
- |
- |
| 5 |
Hb_003697_040 |
0.0731501529 |
- |
- |
hypothetical protein B456_005G209600 [Gossypium raimondii] |
| 6 |
Hb_004696_080 |
0.0757465568 |
- |
- |
PREDICTED: uncharacterized protein At1g01500-like [Populus euphratica] |
| 7 |
Hb_000363_360 |
0.0778470195 |
- |
- |
hypothetical protein PRUPE_ppa009146mg [Prunus persica] |
| 8 |
Hb_005488_200 |
0.078308914 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005305_110 |
0.0831426614 |
- |
- |
PREDICTED: uncharacterized protein LOC105648286 [Jatropha curcas] |
| 10 |
Hb_004126_070 |
0.0838442668 |
desease resistance |
Gene Name: CDC48_2 |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 11 |
Hb_000477_060 |
0.085795215 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 12 |
Hb_019654_030 |
0.0867380774 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 13 |
Hb_000538_070 |
0.0873475373 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 14 |
Hb_000108_190 |
0.0895853883 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Jatropha curcas] |
| 15 |
Hb_086639_030 |
0.092090278 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 16 |
Hb_000023_090 |
0.0926624789 |
- |
- |
mak, putative [Ricinus communis] |
| 17 |
Hb_000016_160 |
0.0930702568 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 18 |
Hb_000985_060 |
0.0937494863 |
- |
- |
PREDICTED: protein transport protein SFT2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000841_020 |
0.0938136369 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 20 |
Hb_002014_020 |
0.093921857 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |