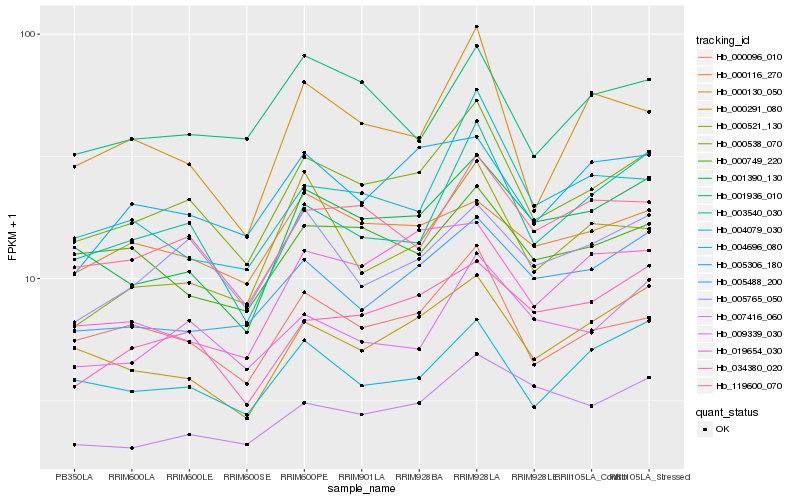
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_070 |
0.0 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 2 |
Hb_000096_010 |
0.0703269378 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 3 |
Hb_119600_070 |
0.0803519381 |
- |
- |
PREDICTED: uncharacterized protein LOC105635190 [Jatropha curcas] |
| 4 |
Hb_001390_130 |
0.0853538803 |
- |
- |
hypothetical protein POPTR_0009s08590g [Populus trichocarpa] |
| 5 |
Hb_000291_080 |
0.0873475373 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 6 |
Hb_009339_030 |
0.0879771222 |
- |
- |
PREDICTED: mitogen-activated protein kinase 9-like [Jatropha curcas] |
| 7 |
Hb_003540_030 |
0.0895515454 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005488_200 |
0.0912331882 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 9 |
Hb_034380_020 |
0.0934699599 |
- |
- |
PREDICTED: phosphoglycerate mutase-like [Jatropha curcas] |
| 10 |
Hb_004696_080 |
0.0944060585 |
- |
- |
PREDICTED: uncharacterized protein At1g01500-like [Populus euphratica] |
| 11 |
Hb_005306_180 |
0.0944814431 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_004079_030 |
0.0945863255 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005765_050 |
0.0947488774 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4-like [Populus euphratica] |
| 14 |
Hb_000130_050 |
0.096378523 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 15 |
Hb_000749_220 |
0.0991831584 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001936_010 |
0.1001339725 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 17 |
Hb_019654_030 |
0.1002627415 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 18 |
Hb_000521_130 |
0.1004762239 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_007416_060 |
0.1005199447 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 18 [Jatropha curcas] |
| 20 |
Hb_000116_270 |
0.1016181384 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |