| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
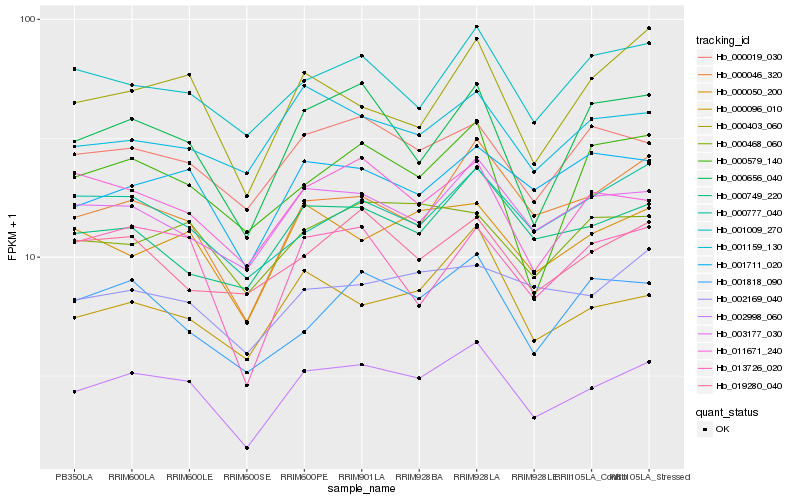
Hb_002998_060 |
0.0 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 2 |
Hb_000046_320 |
0.0824580925 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 3 |
Hb_000656_040 |
0.0841510188 |
- |
- |
PREDICTED: 25.3 kDa vesicle transport protein [Jatropha curcas] |
| 4 |
Hb_003177_030 |
0.0846748034 |
- |
- |
PREDICTED: uncharacterized protein LOC105635357 [Jatropha curcas] |
| 5 |
Hb_000050_200 |
0.0859840845 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 6 |
Hb_000019_030 |
0.0884321628 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 7 |
Hb_000096_010 |
0.0917447704 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 8 |
Hb_001711_020 |
0.0919879035 |
- |
- |
PREDICTED: AP-2 complex subunit mu [Jatropha curcas] |
| 9 |
Hb_000749_220 |
0.0922052622 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000468_060 |
0.0936318532 |
- |
- |
ku P70 DNA helicase, putative [Ricinus communis] |
| 11 |
Hb_011671_240 |
0.0936579387 |
- |
- |
PREDICTED: WD repeat-containing protein WRAP73-like [Jatropha curcas] |
| 12 |
Hb_001009_270 |
0.0946123285 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 6 homolog [Jatropha curcas] |
| 13 |
Hb_001818_090 |
0.0967802939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000403_060 |
0.0982119476 |
- |
- |
PREDICTED: probable ATP synthase 24 kDa subunit, mitochondrial [Vitis vinifera] |
| 15 |
Hb_001159_130 |
0.0986642098 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 16 |
Hb_013726_020 |
0.098925921 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 17 |
Hb_000777_040 |
0.1004036036 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 18 |
Hb_002169_040 |
0.1006940649 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_000579_140 |
0.101348628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_019280_040 |
0.1021253032 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18975, chloroplastic isoform X1 [Jatropha curcas] |