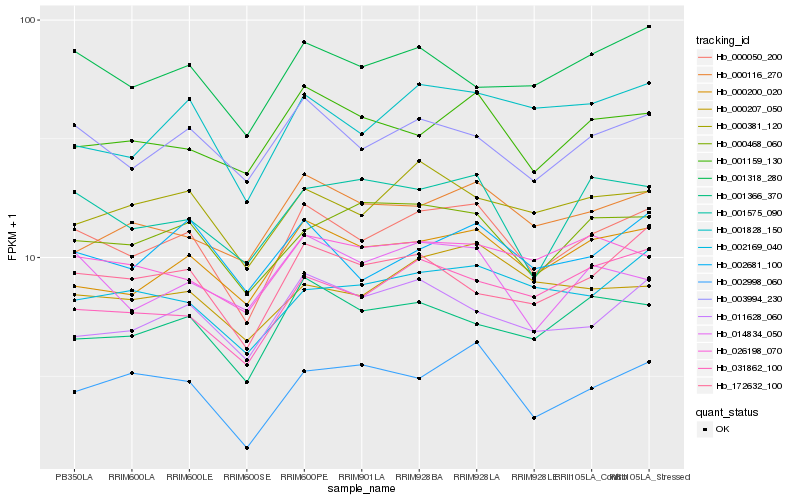
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000050_200 |
0.0 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 2 |
Hb_003994_230 |
0.0682064709 |
- |
- |
7-dehydrocholesterol reductase, putative [Ricinus communis] |
| 3 |
Hb_000200_020 |
0.0753800047 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 4 |
Hb_000468_060 |
0.0808643994 |
- |
- |
ku P70 DNA helicase, putative [Ricinus communis] |
| 5 |
Hb_002681_100 |
0.0818853687 |
- |
- |
PREDICTED: glucuronokinase 1 [Jatropha curcas] |
| 6 |
Hb_001575_090 |
0.0826510412 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 7 |
Hb_001828_150 |
0.0827803494 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001318_280 |
0.0832838532 |
- |
- |
Protein FAM18B, putative [Ricinus communis] |
| 9 |
Hb_001366_370 |
0.0840963697 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001159_130 |
0.0841790932 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 11 |
Hb_000381_120 |
0.084381829 |
- |
- |
PREDICTED: uncharacterized protein LOC105648175 [Jatropha curcas] |
| 12 |
Hb_172632_100 |
0.0858396934 |
- |
- |
PREDICTED: reticulon-like protein B22 [Jatropha curcas] |
| 13 |
Hb_002998_060 |
0.0859840845 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 14 |
Hb_011628_060 |
0.086043952 |
- |
- |
Adenosine 3'-phospho 5'-phosphosulfate transporter, putative [Ricinus communis] |
| 15 |
Hb_014834_050 |
0.0866807681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002169_040 |
0.0872342729 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_031862_100 |
0.0881183968 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_026198_070 |
0.0894081095 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 19 |
Hb_000207_050 |
0.0897808818 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000116_270 |
0.0904589034 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |