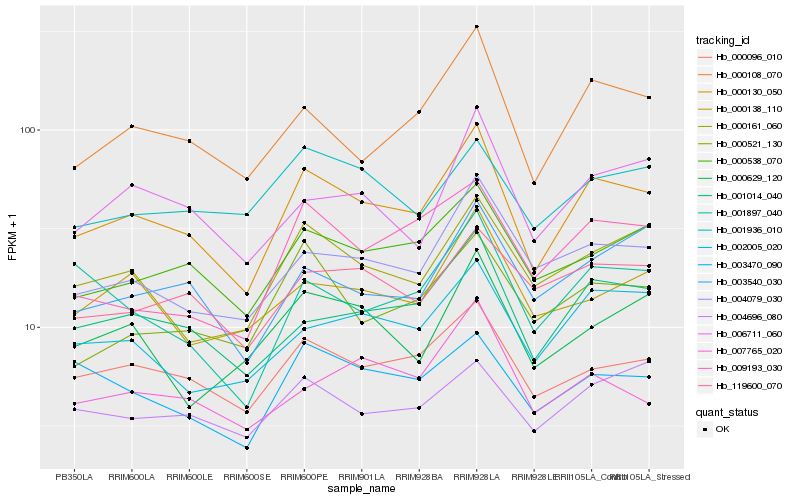
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_050 |
0.0 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 2 |
Hb_000096_010 |
0.0859450434 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 3 |
Hb_001014_040 |
0.0887957021 |
- |
- |
PREDICTED: uncharacterized protein LOC105647308 [Jatropha curcas] |
| 4 |
Hb_000108_070 |
0.0916739526 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 5 |
Hb_000538_070 |
0.096378523 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 6 |
Hb_004079_030 |
0.0973226693 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000161_060 |
0.1021443132 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 [Jatropha curcas] |
| 8 |
Hb_002005_020 |
0.1074504764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006711_060 |
0.1092752739 |
- |
- |
Tubulin beta-1 chain [Morus notabilis] |
| 10 |
Hb_000521_130 |
0.1106194106 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_119600_070 |
0.1122337853 |
- |
- |
PREDICTED: uncharacterized protein LOC105635190 [Jatropha curcas] |
| 12 |
Hb_003540_030 |
0.1122762822 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003470_090 |
0.1128157913 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 14 |
Hb_000138_110 |
0.1150997955 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 15 |
Hb_001936_010 |
0.1164715704 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 16 |
Hb_004696_080 |
0.1165649069 |
- |
- |
PREDICTED: uncharacterized protein At1g01500-like [Populus euphratica] |
| 17 |
Hb_009193_030 |
0.1167796632 |
- |
- |
hypothetical protein POPTR_0010s16620g [Populus trichocarpa] |
| 18 |
Hb_000629_120 |
0.1185869419 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein [Theobroma cacao] |
| 19 |
Hb_001897_040 |
0.1186008889 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_007765_020 |
0.1189872529 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |