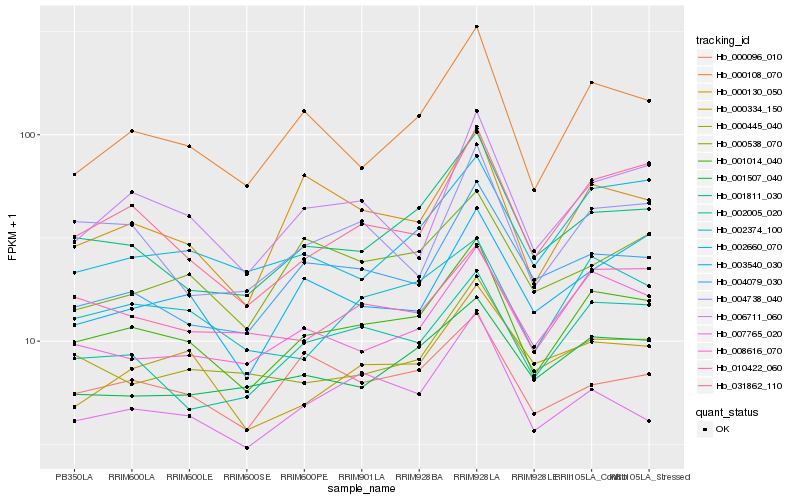
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001014_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647308 [Jatropha curcas] |
| 2 |
Hb_000108_070 |
0.0772003177 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 3 |
Hb_001811_030 |
0.0792849619 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 4 |
Hb_031862_110 |
0.0833718884 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 5 |
Hb_002005_020 |
0.0855503191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002374_100 |
0.0857813143 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 7 |
Hb_000130_050 |
0.0887957021 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 8 |
Hb_008616_070 |
0.0898952912 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 8 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006711_060 |
0.091438338 |
- |
- |
Tubulin beta-1 chain [Morus notabilis] |
| 10 |
Hb_004079_030 |
0.0926108197 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 11 |
Hb_010422_060 |
0.0957771714 |
- |
- |
PREDICTED: GLABRA2 expression modulator [Jatropha curcas] |
| 12 |
Hb_001507_040 |
0.0982762066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004738_040 |
0.099566303 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000445_040 |
0.1002187984 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002660_070 |
0.1006391665 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 16 |
Hb_003540_030 |
0.1007670663 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007765_020 |
0.1007747496 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 18 |
Hb_000096_010 |
0.1009080249 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 19 |
Hb_000334_150 |
0.1015121992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000538_070 |
0.1040996452 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |