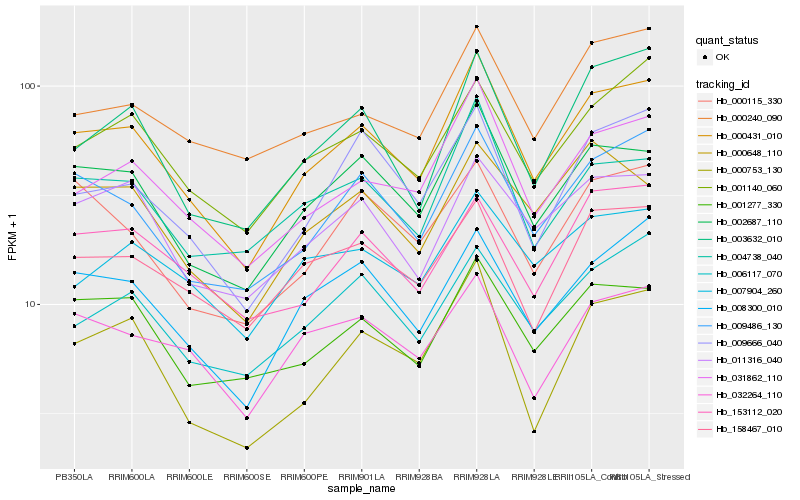
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s05340g [Populus trichocarpa] |
| 2 |
Hb_002687_110 |
0.0592768 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C343.04c isoform X1 [Jatropha curcas] |
| 3 |
Hb_031862_110 |
0.0638552822 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 4 |
Hb_009486_130 |
0.0673232521 |
- |
- |
hypothetical protein CISIN_1g023046mg [Citrus sinensis] |
| 5 |
Hb_008300_010 |
0.0756138336 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 6 |
Hb_001277_330 |
0.0824214704 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 7 |
Hb_001140_060 |
0.0827051295 |
- |
- |
PREDICTED: ADP-ribosylation factor-related protein 1 [Prunus mume] |
| 8 |
Hb_003632_010 |
0.0830613383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009666_040 |
0.0830665878 |
- |
- |
glutathione transferase, putative [Ricinus communis] |
| 10 |
Hb_004738_040 |
0.0849683361 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like isoform X2 [Populus euphratica] |
| 11 |
Hb_006117_070 |
0.0865343779 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 12 |
Hb_011316_040 |
0.0870484618 |
- |
- |
peptidase, putative [Ricinus communis] |
| 13 |
Hb_000753_130 |
0.0886236775 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 14 |
Hb_000240_090 |
0.0895762863 |
- |
- |
Rab5 [Hevea brasiliensis] |
| 15 |
Hb_158467_010 |
0.0902648539 |
- |
- |
transporter, putative [Ricinus communis] |
| 16 |
Hb_032264_110 |
0.0929638201 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 17 |
Hb_153112_020 |
0.0930979961 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 22b-like [Jatropha curcas] |
| 18 |
Hb_000115_330 |
0.0941748868 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 19 |
Hb_000648_110 |
0.0956522001 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007904_260 |
0.0962917651 |
- |
- |
PREDICTED: uncharacterized protein LOC105644715 [Jatropha curcas] |