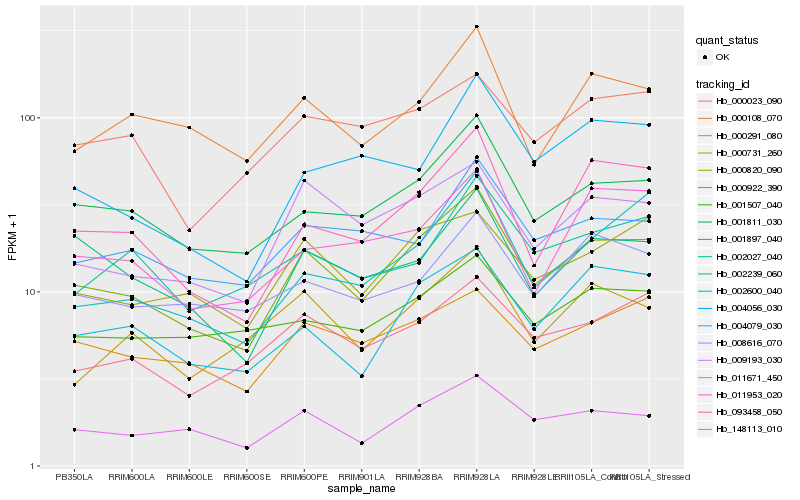
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_390 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 2 |
Hb_011671_450 |
0.0849377552 |
- |
- |
- |
| 3 |
Hb_001811_030 |
0.0883170298 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 4 |
Hb_093458_050 |
0.0988541194 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 5 |
Hb_009193_030 |
0.1006204001 |
- |
- |
hypothetical protein POPTR_0010s16620g [Populus trichocarpa] |
| 6 |
Hb_002600_040 |
0.1037323967 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 7 |
Hb_000820_090 |
0.1048594973 |
- |
- |
- |
| 8 |
Hb_000291_080 |
0.1083554239 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 9 |
Hb_001897_040 |
0.1100449539 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_148113_010 |
0.1102531079 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17C-like isoform X3 [Citrus sinensis] |
| 11 |
Hb_000108_070 |
0.112618931 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 12 |
Hb_004079_030 |
0.117103391 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000023_090 |
0.1202271964 |
- |
- |
mak, putative [Ricinus communis] |
| 14 |
Hb_008616_070 |
0.1203564277 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 8 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000731_260 |
0.1208091394 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 16 |
Hb_002239_060 |
0.1216715149 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 17 |
Hb_011953_020 |
0.1222282695 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 18 |
Hb_004056_030 |
0.1229293553 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 19 |
Hb_001507_040 |
0.1230959702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002027_040 |
0.1243008202 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |