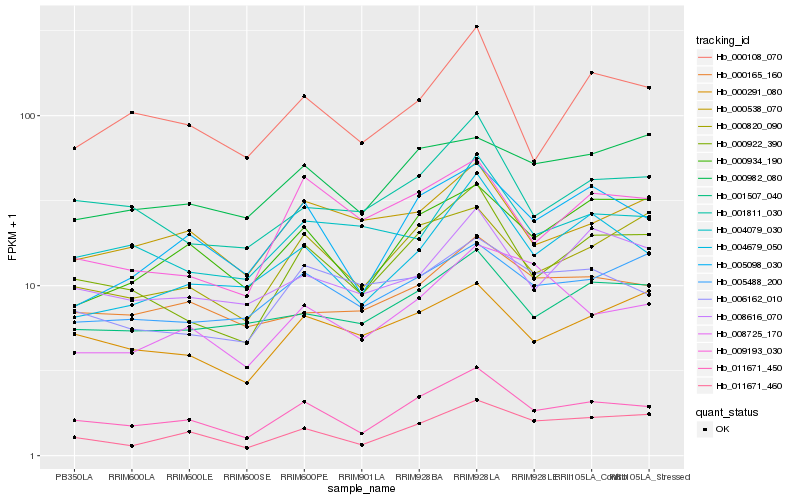
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_450 |
0.0 |
- |
- |
- |
| 2 |
Hb_000922_390 |
0.0849377552 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 3 |
Hb_005098_030 |
0.1041069496 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_011671_460 |
0.10529242 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 5 |
Hb_000820_090 |
0.111756306 |
- |
- |
- |
| 6 |
Hb_004679_050 |
0.1136911788 |
- |
- |
galactono-1,4-lactone dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_001811_030 |
0.1144229569 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 8 |
Hb_000934_190 |
0.1182379032 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 9 |
Hb_000165_160 |
0.1191159827 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |
| 10 |
Hb_001507_040 |
0.119305987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000108_070 |
0.1201853685 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 12 |
Hb_000291_080 |
0.1256219122 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 13 |
Hb_008616_070 |
0.1259033362 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 8 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005488_200 |
0.1261277272 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000538_070 |
0.1269348727 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 16 |
Hb_006162_010 |
0.1274682763 |
- |
- |
CAZy families GH95 protein, partial [uncultured Chitinophaga sp.] |
| 17 |
Hb_009193_030 |
0.1292572377 |
- |
- |
hypothetical protein POPTR_0010s16620g [Populus trichocarpa] |
| 18 |
Hb_008725_170 |
0.1296447238 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 19 |
Hb_000982_080 |
0.1297393416 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 20 |
Hb_004079_030 |
0.1297887528 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |