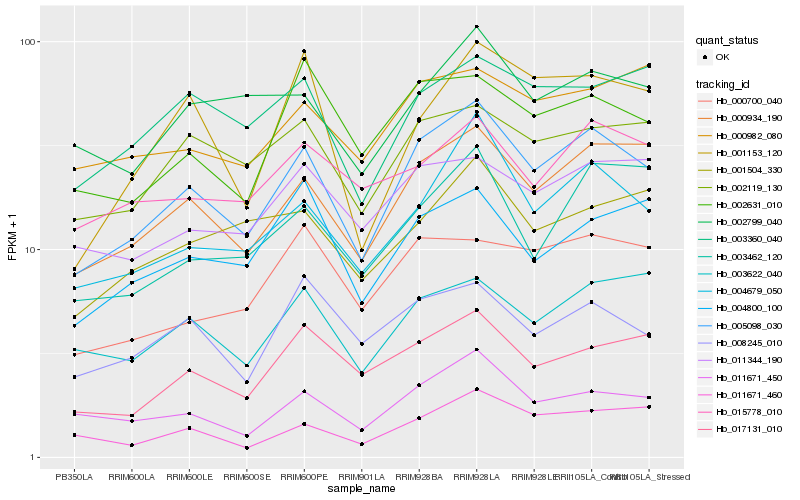
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005098_030 |
0.0 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000934_190 |
0.0782748921 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 3 |
Hb_004679_050 |
0.0960698734 |
- |
- |
galactono-1,4-lactone dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_011671_450 |
0.1041069496 |
- |
- |
- |
| 5 |
Hb_002119_130 |
0.1133729795 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 6 |
Hb_017131_010 |
0.1137954411 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 7 |
Hb_003622_040 |
0.1178081716 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 8 |
Hb_002631_010 |
0.1194574956 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 9 |
Hb_003360_040 |
0.1208304252 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 10 |
Hb_011344_190 |
0.1213340261 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 11 |
Hb_008245_010 |
0.1226056224 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 12 |
Hb_002799_040 |
0.1227171425 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Vitis vinifera] |
| 13 |
Hb_004800_100 |
0.1234774061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003462_120 |
0.1240916884 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 15 |
Hb_000700_040 |
0.1243306779 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |
| 16 |
Hb_011671_460 |
0.1245302064 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 17 |
Hb_015778_010 |
0.1260265028 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 1 [Jatropha curcas] |
| 18 |
Hb_001504_330 |
0.12701115 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 19 |
Hb_000982_080 |
0.1278018609 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 20 |
Hb_001153_120 |
0.1279613881 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |