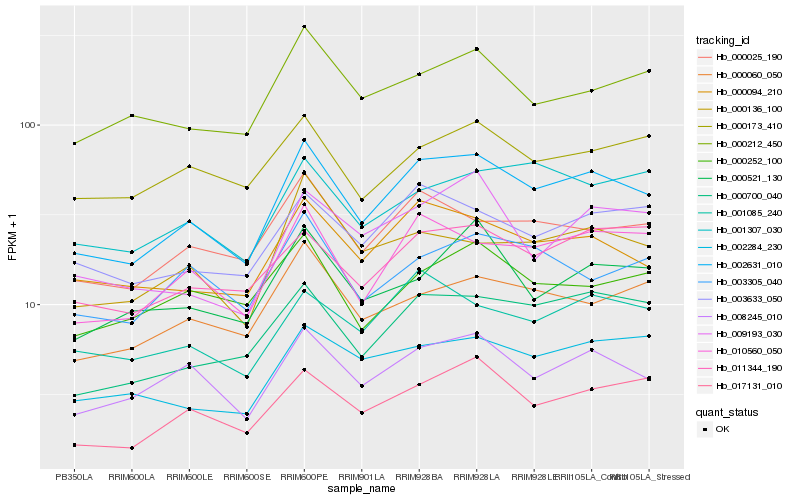
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_010 |
0.0 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 2 |
Hb_008245_010 |
0.0765201354 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 3 |
Hb_010560_050 |
0.0794185081 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 4 |
Hb_000094_210 |
0.0803250369 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 5 |
Hb_000700_040 |
0.0871066492 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |
| 6 |
Hb_000025_190 |
0.0920134054 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 7 |
Hb_017131_010 |
0.0933285751 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 8 |
Hb_003633_050 |
0.0934539061 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 9 |
Hb_001085_240 |
0.0965145651 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011344_190 |
0.098216831 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 11 |
Hb_000060_050 |
0.0989885576 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 12 |
Hb_003305_040 |
0.1030937098 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 13 |
Hb_000136_100 |
0.1035388907 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 14 |
Hb_000212_450 |
0.1037535527 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_002284_230 |
0.1044109104 |
- |
- |
PREDICTED: uncharacterized protein At5g43822 [Jatropha curcas] |
| 16 |
Hb_000173_410 |
0.1050685714 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 17 |
Hb_001307_030 |
0.1055799667 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000252_100 |
0.1063156732 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 19 |
Hb_009193_030 |
0.1082195994 |
- |
- |
hypothetical protein POPTR_0010s16620g [Populus trichocarpa] |
| 20 |
Hb_000521_130 |
0.1087694155 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |