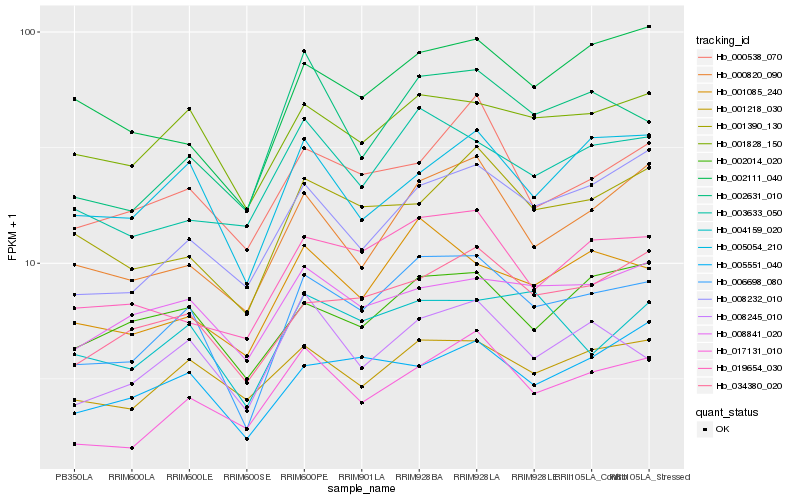
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006698_080 |
0.0 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_034380_020 |
0.1035378493 |
- |
- |
PREDICTED: phosphoglycerate mutase-like [Jatropha curcas] |
| 3 |
Hb_001085_240 |
0.1055931466 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 4 |
Hb_019654_030 |
0.1079393973 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_001828_150 |
0.1079841212 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 6 |
Hb_008245_010 |
0.1082800714 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 7 |
Hb_002631_010 |
0.1099172023 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 8 |
Hb_017131_010 |
0.1110903412 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 9 |
Hb_000820_090 |
0.1120680931 |
- |
- |
- |
| 10 |
Hb_001218_030 |
0.1127775136 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 11 |
Hb_001390_130 |
0.114113203 |
- |
- |
hypothetical protein POPTR_0009s08590g [Populus trichocarpa] |
| 12 |
Hb_008232_010 |
0.1141143078 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 13 |
Hb_002014_020 |
0.1159552394 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004159_020 |
0.1168009193 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 15 |
Hb_005054_210 |
0.1179952057 |
- |
- |
hypothetical protein PRUPE_ppa026456mg [Prunus persica] |
| 16 |
Hb_003633_050 |
0.1192760578 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 17 |
Hb_000538_070 |
0.1196104783 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 18 |
Hb_005551_040 |
0.1205925336 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 19 |
Hb_002111_040 |
0.1211199919 |
- |
- |
hypothetical protein JCGZ_16989 [Jatropha curcas] |
| 20 |
Hb_008841_020 |
0.1225925973 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631075 isoform X1 [Jatropha curcas] |