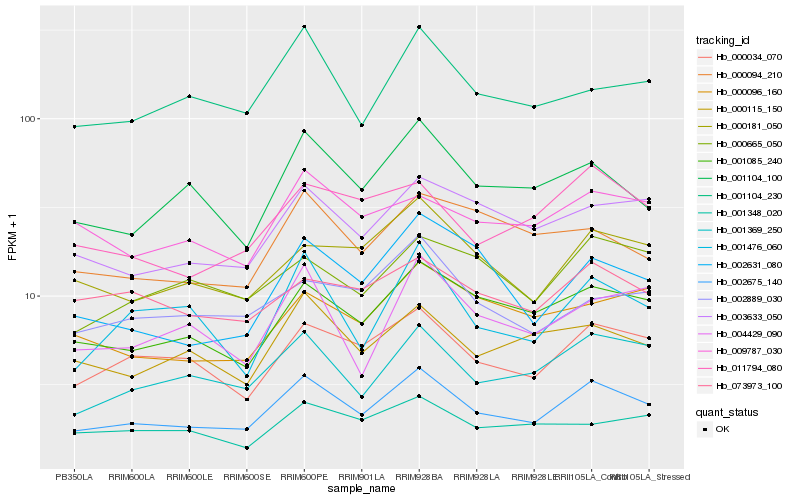
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002675_140 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 2 |
Hb_000034_070 |
0.0828151105 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 3B-like [Jatropha curcas] |
| 3 |
Hb_001085_240 |
0.0859709069 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001369_250 |
0.086024971 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003633_050 |
0.0986991326 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 6 |
Hb_001104_100 |
0.1003961437 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 7 |
Hb_000181_050 |
0.1007641227 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 8 |
Hb_000115_150 |
0.1021065853 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 9 |
Hb_001104_230 |
0.1048421841 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 10 |
Hb_011794_080 |
0.1068009561 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 11 |
Hb_000665_050 |
0.11006468 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 12 |
Hb_001476_060 |
0.1110108087 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 13 |
Hb_002631_080 |
0.1116512969 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_009787_030 |
0.113344718 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 15 |
Hb_004429_090 |
0.1143070699 |
- |
- |
PREDICTED: ras GTPase-activating protein-binding protein 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002889_030 |
0.1161853711 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 17 |
Hb_073973_100 |
0.1166275735 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 18 |
Hb_000096_160 |
0.1168732248 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 19 |
Hb_000094_210 |
0.1169896266 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 20 |
Hb_001348_020 |
0.1176180279 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |