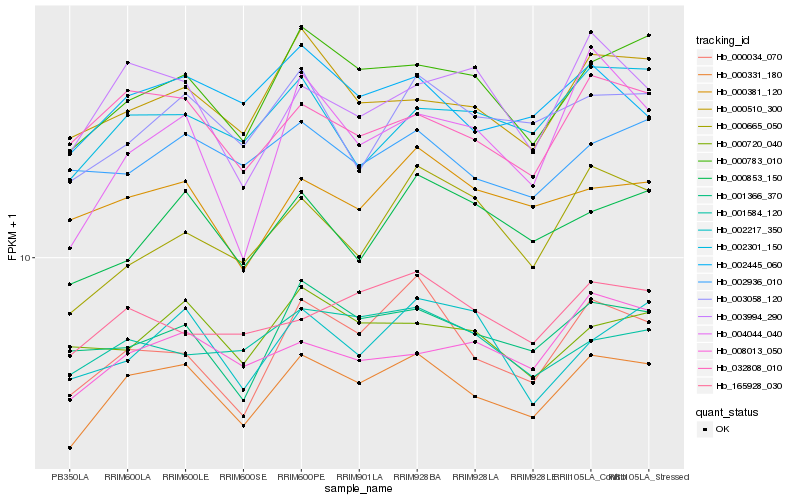
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_180 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640220 [Jatropha curcas] |
| 2 |
Hb_000783_010 |
0.0751517341 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 3 |
Hb_000034_070 |
0.0850596479 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 3B-like [Jatropha curcas] |
| 4 |
Hb_032808_010 |
0.0882590288 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 5 |
Hb_001366_370 |
0.0912611414 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000510_300 |
0.0930326555 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002445_060 |
0.0946575992 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 8 |
Hb_003058_120 |
0.0950719353 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 9 |
Hb_000381_120 |
0.0970400411 |
- |
- |
PREDICTED: uncharacterized protein LOC105648175 [Jatropha curcas] |
| 10 |
Hb_000853_150 |
0.0973484782 |
- |
- |
Fumarase 1 isoform 2 [Theobroma cacao] |
| 11 |
Hb_002301_150 |
0.0976921696 |
- |
- |
Drought-responsive family protein [Theobroma cacao] |
| 12 |
Hb_003994_290 |
0.101044944 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase GAPCP2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_008013_050 |
0.1011470569 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 14 |
Hb_165928_030 |
0.1015376134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000720_040 |
0.1033957449 |
- |
- |
PREDICTED: replication factor C subunit 2 [Jatropha curcas] |
| 16 |
Hb_002936_010 |
0.1038769628 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 17 |
Hb_004044_040 |
0.1048355469 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 18 |
Hb_001584_120 |
0.105044195 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase 3 [Jatropha curcas] |
| 19 |
Hb_000665_050 |
0.1052169549 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 20 |
Hb_002217_350 |
0.1052934201 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |