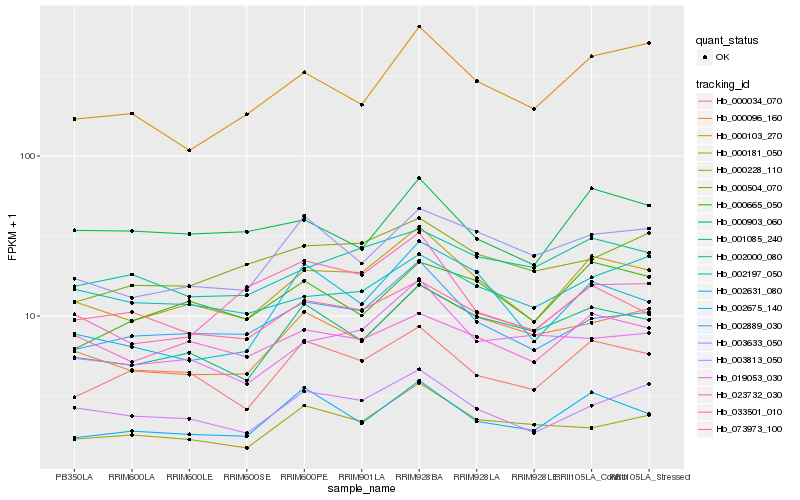
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 2 |
Hb_002889_030 |
0.0796477879 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 3 |
Hb_003813_050 |
0.0893154618 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001085_240 |
0.0939952687 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000034_070 |
0.0940771318 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 3B-like [Jatropha curcas] |
| 6 |
Hb_000096_160 |
0.1005628589 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 7 |
Hb_002675_140 |
0.1007641227 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 8 |
Hb_023732_030 |
0.1012295802 |
- |
- |
PREDICTED: proteinaceous RNase P 2-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_002000_080 |
0.1024983828 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_073973_100 |
0.1036371103 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 11 |
Hb_019053_030 |
0.1039600327 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 12 |
Hb_003633_050 |
0.1068080542 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 13 |
Hb_000103_270 |
0.1071129665 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_000504_070 |
0.110550071 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_000228_110 |
0.1105818515 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000665_050 |
0.110677959 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 17 |
Hb_002197_050 |
0.1118249965 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |
| 18 |
Hb_000903_060 |
0.1126820146 |
- |
- |
PREDICTED: ras-related protein RABA5e [Jatropha curcas] |
| 19 |
Hb_002631_080 |
0.1127271203 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_033501_010 |
0.1132151166 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |