| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
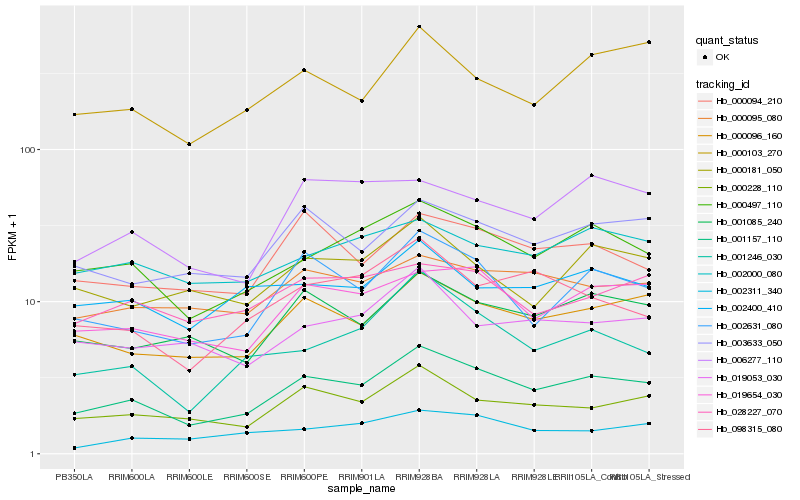
Hb_001157_110 |
0.0 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 2 |
Hb_000497_110 |
0.0836226292 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 3 |
Hb_000096_160 |
0.0857167288 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 4 |
Hb_019654_030 |
0.0923213999 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_002631_080 |
0.0941267956 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000228_110 |
0.0959666891 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001085_240 |
0.0965198669 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003633_050 |
0.1039458943 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 9 |
Hb_000094_210 |
0.1128330171 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 10 |
Hb_000103_270 |
0.1137536211 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_000095_080 |
0.1140079108 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 12 |
Hb_001246_030 |
0.1157929125 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 13 |
Hb_002000_080 |
0.1176430311 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002400_410 |
0.1179169116 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 15 |
Hb_019053_030 |
0.1185539974 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 16 |
Hb_098315_080 |
0.1187374597 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_006277_110 |
0.1193530751 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000181_050 |
0.1202054006 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 19 |
Hb_028227_070 |
0.1214551775 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002311_340 |
0.1220513316 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |