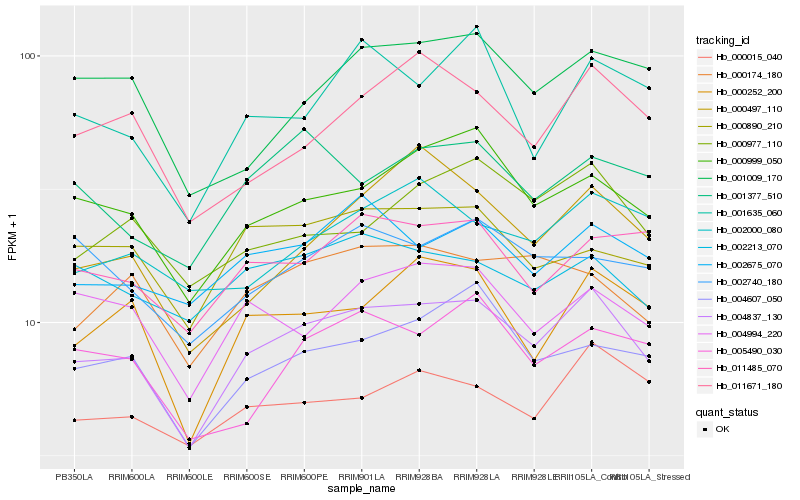
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_130 |
0.0 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 2 |
Hb_000999_050 |
0.0784103269 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000252_200 |
0.0864787048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004607_050 |
0.0891546344 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_002675_070 |
0.0918191616 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 6 |
Hb_000174_180 |
0.09300056 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_011485_070 |
0.0936913344 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 8 |
Hb_004994_220 |
0.0938978533 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_011671_180 |
0.0942097564 |
- |
- |
PREDICTED: probable quinone oxidoreductase [Jatropha curcas] |
| 10 |
Hb_000015_040 |
0.0948641367 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000977_110 |
0.0949051339 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000890_210 |
0.0965397209 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 isoform X1 [Eucalyptus grandis] |
| 13 |
Hb_005490_030 |
0.0976050144 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2 isoform X3 [Jatropha curcas] |
| 14 |
Hb_001377_510 |
0.0993281846 |
- |
- |
ormdl, putative [Ricinus communis] |
| 15 |
Hb_002213_070 |
0.0994315988 |
- |
- |
epsilon-adaptin family protein [Populus trichocarpa] |
| 16 |
Hb_001635_060 |
0.1000110359 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 17 |
Hb_002000_080 |
0.100153248 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000497_110 |
0.1004857014 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 19 |
Hb_001009_170 |
0.100570159 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_002740_180 |
0.1015029222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |