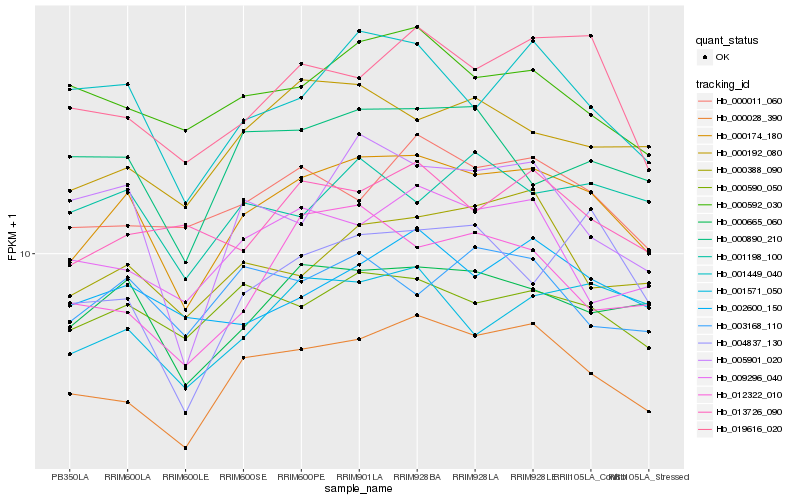
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000174_180 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000028_390 |
0.063914836 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 3 |
Hb_000665_060 |
0.0649873106 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 4 |
Hb_001449_040 |
0.0811490748 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000590_050 |
0.0823422915 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 6 |
Hb_000192_080 |
0.0829687399 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_019616_020 |
0.0847002998 |
- |
- |
PREDICTED: alanine--tRNA ligase [Jatropha curcas] |
| 8 |
Hb_001571_050 |
0.0853911717 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |
| 9 |
Hb_000388_090 |
0.0854930122 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_000011_060 |
0.0879392788 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002600_150 |
0.0880221732 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000890_210 |
0.089159777 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 isoform X1 [Eucalyptus grandis] |
| 13 |
Hb_012322_010 |
0.0908544613 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000592_030 |
0.0911604561 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 15 |
Hb_013726_090 |
0.0921083211 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 16 |
Hb_003168_110 |
0.0925984387 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 17 |
Hb_004837_130 |
0.09300056 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 18 |
Hb_009296_040 |
0.0931159847 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 19 |
Hb_005901_020 |
0.0933083703 |
- |
- |
PREDICTED: elongation factor Tu GTP-binding domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_001198_100 |
0.0944771327 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7-like [Jatropha curcas] |