| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
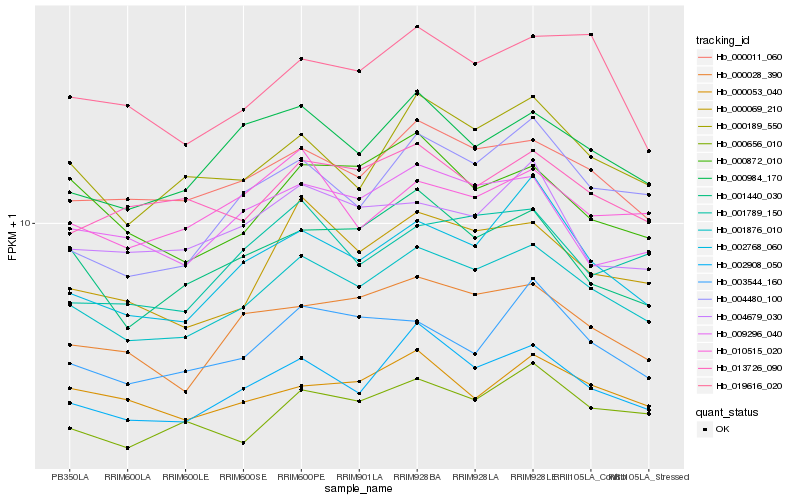
Hb_001876_010 |
0.0 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000011_060 |
0.0513445058 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002908_050 |
0.0517136712 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 4 |
Hb_000189_550 |
0.0585651961 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 5 |
Hb_000069_210 |
0.0608271479 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 6 |
Hb_000028_390 |
0.0626473423 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 7 |
Hb_000984_170 |
0.0633739265 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 8 |
Hb_002768_060 |
0.0637067632 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000656_010 |
0.0661522809 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001440_030 |
0.068356153 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 11 |
Hb_019616_020 |
0.069542885 |
- |
- |
PREDICTED: alanine--tRNA ligase [Jatropha curcas] |
| 12 |
Hb_003544_160 |
0.0702644376 |
- |
- |
- |
| 13 |
Hb_009296_040 |
0.0704956305 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 14 |
Hb_000872_010 |
0.0720581186 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 15 |
Hb_000053_040 |
0.0723083621 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 16 |
Hb_013726_090 |
0.0727515929 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 17 |
Hb_004480_100 |
0.0742962622 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 18 |
Hb_001789_150 |
0.0748130394 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_010515_020 |
0.0748146029 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 20 |
Hb_004679_030 |
0.0755377611 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |