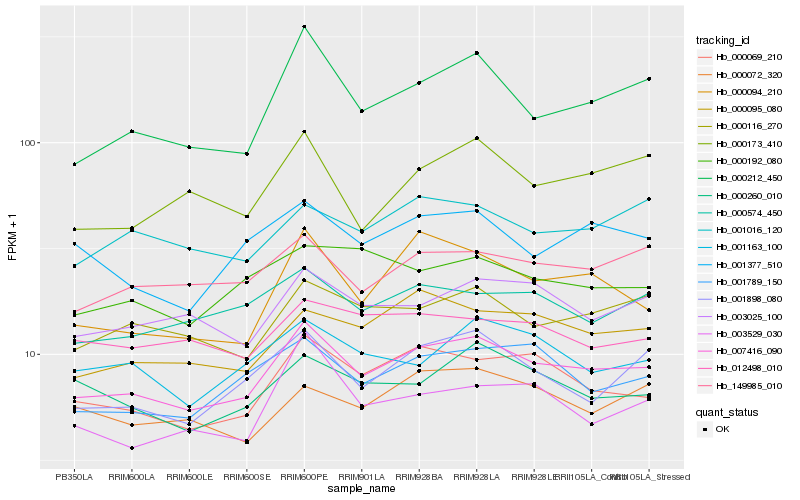
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_090 |
0.0 |
- |
- |
UDP-sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_000069_210 |
0.0642692512 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 3 |
Hb_001898_080 |
0.0728491581 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000116_270 |
0.0733510367 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |
| 5 |
Hb_001789_150 |
0.0739877986 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_149985_010 |
0.0742868224 |
- |
- |
PREDICTED: transcription initiation factor IIB-2 [Cucumis sativus] |
| 7 |
Hb_000095_080 |
0.0743234023 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 8 |
Hb_000212_450 |
0.0756801356 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 9 |
Hb_003025_100 |
0.075873516 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000574_450 |
0.0785048183 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 11 |
Hb_001377_510 |
0.0801246021 |
- |
- |
ormdl, putative [Ricinus communis] |
| 12 |
Hb_000260_010 |
0.0805975884 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 13 |
Hb_000094_210 |
0.0816319619 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 14 |
Hb_001163_100 |
0.0816663861 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001016_120 |
0.0840632151 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000072_320 |
0.0858672863 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 17 |
Hb_000173_410 |
0.0863151324 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 18 |
Hb_012498_010 |
0.0879545096 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 19 |
Hb_003529_030 |
0.0879706743 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 20 |
Hb_000192_080 |
0.0884202015 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK3 isoform X1 [Jatropha curcas] |