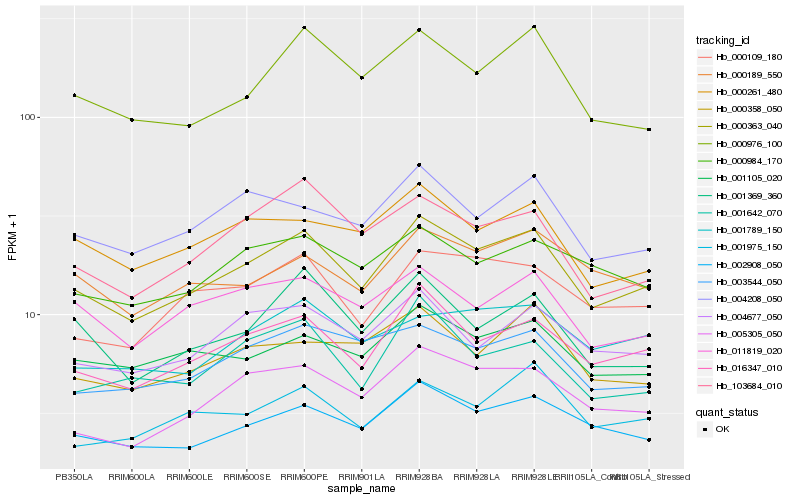
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_040 |
0.0 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 2 |
Hb_016347_010 |
0.0563177875 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 3 |
Hb_002908_050 |
0.0657562486 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 4 |
Hb_005305_050 |
0.0730396983 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 5 |
Hb_004677_050 |
0.0766603392 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000109_180 |
0.0770617885 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 7 |
Hb_000976_100 |
0.0780869268 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 8 |
Hb_004208_050 |
0.0785009847 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 9 |
Hb_001105_020 |
0.0790336615 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 10 |
Hb_000261_480 |
0.0791039114 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 11 |
Hb_103684_010 |
0.0805527505 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 12 |
Hb_011819_020 |
0.0809031319 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003544_050 |
0.0816347595 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000984_170 |
0.0823758041 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 15 |
Hb_000189_550 |
0.0840467645 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 16 |
Hb_001975_150 |
0.0844799986 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 17 |
Hb_001369_360 |
0.0851665381 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 18 |
Hb_001789_150 |
0.0859872048 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000358_050 |
0.0865948964 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 20 |
Hb_001642_070 |
0.0867061271 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |