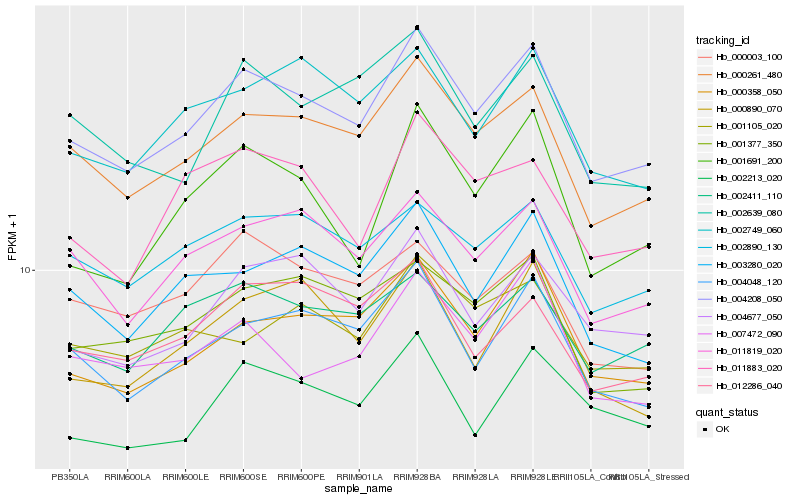
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004208_050 |
0.0 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 2 |
Hb_000261_480 |
0.0390134628 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 3 |
Hb_004048_120 |
0.0571358921 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000358_050 |
0.0580386886 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 5 |
Hb_011819_020 |
0.0586070221 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002890_130 |
0.0634314094 |
- |
- |
tip120, putative [Ricinus communis] |
| 7 |
Hb_002411_110 |
0.0641702325 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 8 |
Hb_003280_020 |
0.0644171858 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004677_050 |
0.0645916228 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001377_350 |
0.0668067044 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 11 |
Hb_002749_060 |
0.0680719721 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 12 |
Hb_007472_090 |
0.0711795447 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_001691_200 |
0.0713338758 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 14 |
Hb_002639_080 |
0.0717163676 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011883_020 |
0.0719960042 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 16 |
Hb_012286_040 |
0.0727387525 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 17 |
Hb_002213_020 |
0.0731592279 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000003_100 |
0.0732717148 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 19 |
Hb_000890_070 |
0.0734550998 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001105_020 |
0.0735969884 |
- |
- |
expressed protein, putative [Ricinus communis] |