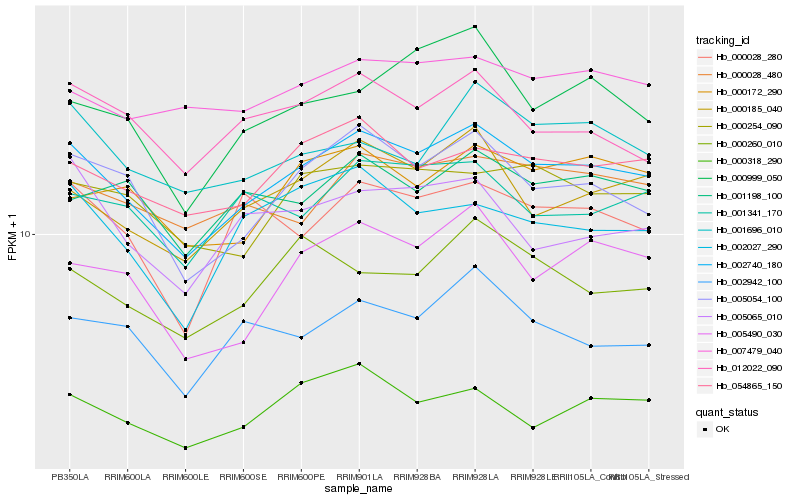
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002740_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005054_100 |
0.059519477 |
- |
- |
PREDICTED: uncharacterized protein At2g39920 [Jatropha curcas] |
| 3 |
Hb_000185_040 |
0.0632001927 |
- |
- |
PREDICTED: uncharacterized protein LOC103343434 [Prunus mume] |
| 4 |
Hb_054865_150 |
0.0660288492 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein Q [Jatropha curcas] |
| 5 |
Hb_000254_090 |
0.0719684281 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 6 |
Hb_000318_290 |
0.0725099301 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000028_480 |
0.0727592955 |
- |
- |
PREDICTED: putative 3,4-dihydroxy-2-butanone kinase [Jatropha curcas] |
| 8 |
Hb_012022_090 |
0.0741281552 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Gossypium raimondii] |
| 9 |
Hb_005490_030 |
0.0744583499 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2 isoform X3 [Jatropha curcas] |
| 10 |
Hb_002027_290 |
0.0754871415 |
- |
- |
PREDICTED: NPL4-like protein 1 [Jatropha curcas] |
| 11 |
Hb_001341_170 |
0.0760735928 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 12 |
Hb_002942_100 |
0.0763666192 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61990, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000172_290 |
0.0765259964 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 14 |
Hb_001198_100 |
0.0771440111 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7-like [Jatropha curcas] |
| 15 |
Hb_000028_280 |
0.0776909398 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 16 |
Hb_007479_040 |
0.0777502494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001696_010 |
0.0788431749 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_000260_010 |
0.0806546297 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 19 |
Hb_005065_010 |
0.080673467 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI11 [Jatropha curcas] |
| 20 |
Hb_000999_050 |
0.0813482213 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |